Volume 12, Issue 3 (September 2025)
J. Food Qual. Hazards Control 2025, 12(3): 171-182 |
Back to browse issues page
Ethics code: Not applicable.
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Onipede G, Omodara E, Sanni A. Antifungal Activity of Lactobacillus plantarum and Lactobacillus fermentum Isolated from Agadagidi and Ogi against Spoilage Fungi of Plantain. J. Food Qual. Hazards Control 2025; 12 (3) :171-182
URL: http://jfqhc.ssu.ac.ir/article-1-1244-en.html
URL: http://jfqhc.ssu.ac.ir/article-1-1244-en.html
Department of Microbiology, Federal University of Health Sciences, Ila-Orangun, Nigeria , gbemisola.onipede@fuhsi.edu.ng
Keywords: Antifungal Agents, Aspergillus flavus, Fermented Foods, Lactobacillus plantarum, Penicillium, Public Health
Full-Text [PDF 553 kb]
(233 Downloads)
| Abstract (HTML) (355 Views)
Full-Text: (8 Views)
Antifungal Activity of Lactobacillus plantarum and Lactobacillus fermentum Isolated from Agadagidi and Ogi against Spoilage Fungi of Plantain
G.O. Onipede ** , E.K. Omodara 2, A.I. Sanni 2
1. Department of Microbiology, Federal University of Health Sciences, Ila-Orangun, Nigeria
2. Department of Microbiology, University of Ibadan, Ibadan, Nigeria
HIGHLIGHTS
Table 1: Total counts of Lactic Acid Bacteria (LAB) and fungi in ogi and agadagidi
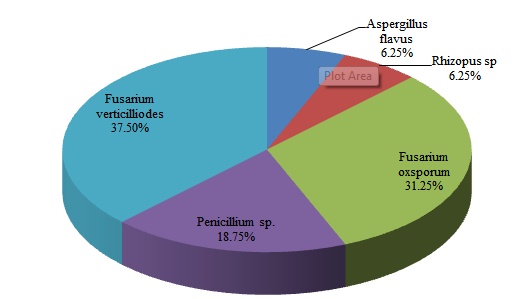 Figure 1: Frequency of occurrence (%) of fungal isolates from plantain
Figure 1: Frequency of occurrence (%) of fungal isolates from plantain
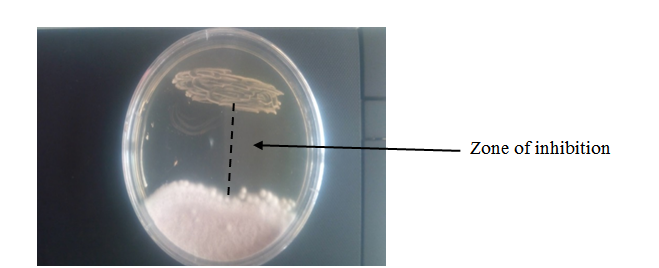
Figure 2: Antifungal activity of Lactobacillus fermentum 5AG8 against the growth of Fusarium oxysporum using agar overlay method
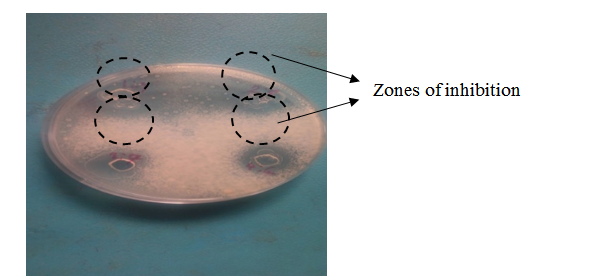
Figure 3: Zones of inhibition produced by the Cell-Free Supernatants (CFS) of Lactic Acid Bacteria (LAB) isolate 5AG8 against Aspergillus flavus and Fusarium oxysporum
Table 5: Phenotypic characteristics of Lactic Acid Bacteria (LAB) Isolates with antifungal activity
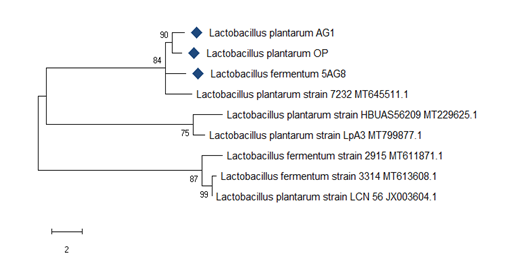
Figure 4: Evolutionary analysis constructed using the maximum likelihood technique
G.O. Onipede **
1. Department of Microbiology, Federal University of Health Sciences, Ila-Orangun, Nigeria
2. Department of Microbiology, University of Ibadan, Ibadan, Nigeria
- Lactobacillus fermentum 5AG8 exhibited the strongest antifungal potential against a wide range of the moulds with highest inhibition produced against Fusarium oxysporum.
- Lactobacillus plantarum AG1, L. plantarum OP, and L. fermentum 5AG8 proved to possess the potential to inhibit spoilage fungi in plantain.
- Safety assessment tests of the LAB isolates revealed that all the isolates are safe to be used in food matrices.
| Article type Original article |
ABSTRACT Background: There has been an increase in public health concern over post-harvest spoilage fungi in plantain. This study aimed to explore the role of Lactic Acid Bacteria (LAB) from fermented foods against spoilage fungi of plantain. Methods: Seventy-five LAB were isolated from agadagidi and ogi between February and April 2021. Cell-Free Supernatants (CFS) of the broth cultures were obtained by centrifugation. Spoilage fungi were obtained from plantain with visible mould growth. Initial screening of the LAB isolates for antifungal activities was carried out using the agar overlay method. The agar well diffusion method was used to assay the inhibitory spectrum of the CFS. Data obtained for the inhibition diameters were analysed using descriptive statistics (IBM SPSS 22). LAB isolates with the highest antifungal activity were assessed for their safety via the DNase, gelatinase, and haemolysis tests, and were identified by 16S rRNA sequencing. Results: Twenty-six LAB isolates exhibiting antifungal activity were obtained from the samples. The isolated spoilage fungi included Aspergillus flavus, Fusarium oxysporum, Fusarium verticilliodes, Penicillium sp, and Rhizopus sp. Nine of the 26 LAB isolates produced inhibition zones >14 mm in diameter. The CFS of isolate 5AG8 had antifungal activity against the spores of A. flavus, F. oxysporum, and Penicillium sp. The CFS of isolate AG1 inhibited the mycelial growth of F. verticilliodes and Penicillium sp, while the CFS of isolate OP was active against the spores of A. flavus and Penicillium sp. None of the isolates tested positive in the safety assessment tests. The isolates were identified as Lactobacillus plantarum OP, L. plantarum AG1, and Lactobacillus fermentum 5AG8. Conclusion: This study revealed that LAB strains from agadagidi and ogi are safe for use in food matrices and their metabolites can be used in the control of spoilage fungi in plantain. © 2025, Shahid Sadoughi University of Medical Sciences. This is an open access article under the Creative Commons Attribution 4.0 International License. |
|
| Keywords Antifungal Agents Aspergillus flavus Fermented Foods Lactobacillus plantarum Penicillium Public Health |
||
| Article history Received: 11 Aug 2024 Revised: 3 Feb 2025 Accepted: 25 Aug 2025 |
||
| Abbreviations CFU=Colony Forming Unit CFS=Cell-Free Supernatants LAB=Lactic Acid Bacteria MRS=De Mann Rogosa Sharpe Agar PCR=Polymerase Chain Reaction PDA=Potato Dextrose Agar |
To cite: Onipede G.O., Omodara E.K., Sanni A.I. (2025). Antifungal activity of Lactobacillus plantarum and Lactobacillus fermentum isolated from agadagidi and ogi against spoilage fungi of plantain. Journal of Food Quality and Hazards Control. 12: 171-182.
Introduction
Introduction
Plantain belongs to the family Musaceae, which is one of the six families within the order Zingibarales. It is one of the known sources of carbohydrate to over 70 million people in West and Central Africa (Abiodun-Solanke and Falade, 2010; Robinson et al., 1992). Plantain is an important source of minerals and vitamins, such as calcium, potassium, iron, manganese, vitamin A, vitamin B6, vitamin C, and dietary fibre. Thus, it plays a vital role in the human nutrition within these regions (Honfo et al., 2007). According to International Institute of Tropical Agriculture (IITA, 2000), it provides more than 25% of the daily intake of carbohydrates and 10% of calories, for over 70 million people in Sub-Sahara Africa. Plantain is known for its post-harvest spoilage. It has been reported that about 30% of fruits and vegetables produced yearly are rendered unfit for consumption due to spoilage (Chavan et al., 2023; Udoh et al., 2015). Storage and maintenance in fresh state is highly difficult to achieve (Rose-Monde et al., 2013). Plantain is sometimes processed into a more stable and convenient form to maintain good keeping quality over extended periods due to its highly perishable nature.
One of the major problems associated with post-harvest storage of foods and feeds is spoilage and poisoning by fungi such as Aspergillus spp, Fusarium spp., Penicillium spp. (Abdu et al., 2024). The predominance of fungi as the organism responsible for the spoilage of some edible fruits and vegetables has been recorded in various regions of the world including Nigeria (Ramudingana et al., 2024; Udoh et al., 2015). The post-harvest spoilage is aided by direct contact with contaminated soil, dust, water, and poor handling during harvesting and processing (Eni et al., 2010). Most fungi linked with the spoilage of plantain fruit are found to be pathogenic to the plantain tree, causing general soft rot (Chuku, 2009). Aspergillus niger causes deep brown to black rot; Fusarium moniliforme causes thick whitish rot; Penicillium frequentans causes green rot; while Rhizopus stolonifer causes pinkish white rot. Cryptococcus neoformans was reportedly encountered when the plantain is fully ripened or spoilt (Chuku, 2009). Microorganisms induced fruit spoilage by secreting the extracellular lytic enzymes that is used to degrade the fruits cell wall polymers and subsequently releasing cell contents and water. Fungi produced the extracellular enzymes (pectinases and hemicellulases) in large quantities, which are required for fungal spoilage processes (Miedes and Lorences, 2004).
Lactic Acid Bacteria (LAB) are characterized as rod or coccal-shaped bacteria that are gram-positive, catalase-negative, with tolerance to low pH (Bin Masalam et al., 2018). Apart from providing health benefits to human (Begley et al., 2006; Rios Covian et al., 2016; Zanotti et al., 2015), LAB are an important source of natural preservatives in food. LAB are known for production of important metabolites in traditional fermented foods and other conventional foods. The metabolites are proven to be effective in elongating the shelf-life of fermented food and feeds (Shehata et al., 2019). Metabolites of natural origin are well recommended for drug and preservatives worldwide. However, their application in food preservation is facing serious challenges due to food spoilage resulting from fungi infections (Rouse et al., 2008).
Post-harvest spoilage of plantain has been well investigated by several researchers, indicating an increasing concern for the development of food grade biopreservatives aimed at ensuring good quality over the period of storage leading to improved market value and extended shelf life of plantain (Mafe et al., 2024; Ofosu et al., 2023). Investigation of the biopreservative potential of LAB has shown that many antimicrobial compounds identified in several species of LAB have antifungal activity (Zapaśnik et al., 2022). The inhibitory influence of these antifungal compounds against mould species, associated with post-harvest spoilage of plantain, is not a doubt. The antifungal compounds produced by LAB have been known to contain peptides with considerable inhibitory effects on yeasts and filamentous fungi such as Aspergillus flavus (Huan et al., 2020). These peptides are characterized by their stability in both acidic and basic environments and at high temperatures, which make them more suitable than chemical preservatives in a wide range of food products (Salas et al., 2017). Typical among these antimicrobial peptides are bacteriocins produced by gram-positive and gram-negative bacteria, with Lactobacillus, Pediococcus, and Leuconostoc species reported to be the most commonly associated with antifungal bacteriocins (Salas et al., 2017, Silva et al., 2018).
Other metabolites also produced by LAB include organic acids (acetic, propionic, lactic, and phenyllactic acids), fatty acids, carboxylic acids, reuterin, and nisin (Crowley et al., 2013, Liu et al., 2021). Despite the successes recorded for these natural antimicrobials in food preservation, their effectiveness must be carefully considered based on observed drawbacks including adsorption of the peptides to food components, poor solubility, enzymatic degradation, irregular distribution in the food matrix, and interactions between these natural additives and food components, food characteristics (pH, texture, etc.), and processing techniques. Optimization of the application conditions can help solve many of these drawbacks (Liu et al., 2021).
One of the strategies developed in recent years to reduce post-harvest losses of plantain in African is recipe development (Strano et al., 2022, Vida et al., 2024). These strategies, which include general recipes, site recipes, master recipes, and control recipes, were developed to minimised post-harvest losses especially in unripe plantain. However, only a limited number of recipes can be developed for unripe plantains, hence significant losses are still inevitable as most of the plantains become ripen under storage (Vida et al., 2024).
In order to decrease the post-harvest spoilage of plantain and enhance their safety, LAB based antifungal metabolites should be developed and effectively applied. Therefore, the present study was focused on evaluating the antifungal activities of LAB and their metabolites against post-harvest spoilage fungi of plantain with the aim of obtaining an alternative biopreservative approach for plantain.
Materials and methods
Materials
The materials used in this study included De Mann Rogosa Sharpe medium (MRS) (Himedia® M641), Potato Dextrose Agar (PDA) (Himedia® MH096), DNase agar (Thermo Scientific™ PO0128A), gelatinase agar (Merck DEV 1106850500), and Zymo Research Quick-DNA™ Miniprep Plus Kit (Zymo Research NC0933233).
Media preparation
MRS agar and PDA used for the isolation of LAB and moulds, respectively, were prepared as instructed by the manufacturer. The media were dissolved and sterilized at 121 oC for 15 min using the autoclave. Sterile media were cooled to about 45-50 oC before pouring into sterile Petri dishes.
Sample collection
A total of sixty samples of agadagidi and ogi were collected aseptically in sterile air tight containers with well fitted cover lids and transported to the laboratory for further analysis. Bunches of freshly harvested plantain with no observable sign of spoilage were collected from Ibadan metropolis, Nigeria, between February and April 2021, for this study, for this study. The fingers were sorted and cleaned under running tap water. These samples were surface sterilized with 1% sodium hypochlorite and stored in clean sterile containers at room temperature and relative humidity for 7-10 days until physical signs of mould spoilage appeared (Chuku, 2009).
One of the major problems associated with post-harvest storage of foods and feeds is spoilage and poisoning by fungi such as Aspergillus spp, Fusarium spp., Penicillium spp. (Abdu et al., 2024). The predominance of fungi as the organism responsible for the spoilage of some edible fruits and vegetables has been recorded in various regions of the world including Nigeria (Ramudingana et al., 2024; Udoh et al., 2015). The post-harvest spoilage is aided by direct contact with contaminated soil, dust, water, and poor handling during harvesting and processing (Eni et al., 2010). Most fungi linked with the spoilage of plantain fruit are found to be pathogenic to the plantain tree, causing general soft rot (Chuku, 2009). Aspergillus niger causes deep brown to black rot; Fusarium moniliforme causes thick whitish rot; Penicillium frequentans causes green rot; while Rhizopus stolonifer causes pinkish white rot. Cryptococcus neoformans was reportedly encountered when the plantain is fully ripened or spoilt (Chuku, 2009). Microorganisms induced fruit spoilage by secreting the extracellular lytic enzymes that is used to degrade the fruits cell wall polymers and subsequently releasing cell contents and water. Fungi produced the extracellular enzymes (pectinases and hemicellulases) in large quantities, which are required for fungal spoilage processes (Miedes and Lorences, 2004).
Lactic Acid Bacteria (LAB) are characterized as rod or coccal-shaped bacteria that are gram-positive, catalase-negative, with tolerance to low pH (Bin Masalam et al., 2018). Apart from providing health benefits to human (Begley et al., 2006; Rios Covian et al., 2016; Zanotti et al., 2015), LAB are an important source of natural preservatives in food. LAB are known for production of important metabolites in traditional fermented foods and other conventional foods. The metabolites are proven to be effective in elongating the shelf-life of fermented food and feeds (Shehata et al., 2019). Metabolites of natural origin are well recommended for drug and preservatives worldwide. However, their application in food preservation is facing serious challenges due to food spoilage resulting from fungi infections (Rouse et al., 2008).
Post-harvest spoilage of plantain has been well investigated by several researchers, indicating an increasing concern for the development of food grade biopreservatives aimed at ensuring good quality over the period of storage leading to improved market value and extended shelf life of plantain (Mafe et al., 2024; Ofosu et al., 2023). Investigation of the biopreservative potential of LAB has shown that many antimicrobial compounds identified in several species of LAB have antifungal activity (Zapaśnik et al., 2022). The inhibitory influence of these antifungal compounds against mould species, associated with post-harvest spoilage of plantain, is not a doubt. The antifungal compounds produced by LAB have been known to contain peptides with considerable inhibitory effects on yeasts and filamentous fungi such as Aspergillus flavus (Huan et al., 2020). These peptides are characterized by their stability in both acidic and basic environments and at high temperatures, which make them more suitable than chemical preservatives in a wide range of food products (Salas et al., 2017). Typical among these antimicrobial peptides are bacteriocins produced by gram-positive and gram-negative bacteria, with Lactobacillus, Pediococcus, and Leuconostoc species reported to be the most commonly associated with antifungal bacteriocins (Salas et al., 2017, Silva et al., 2018).
Other metabolites also produced by LAB include organic acids (acetic, propionic, lactic, and phenyllactic acids), fatty acids, carboxylic acids, reuterin, and nisin (Crowley et al., 2013, Liu et al., 2021). Despite the successes recorded for these natural antimicrobials in food preservation, their effectiveness must be carefully considered based on observed drawbacks including adsorption of the peptides to food components, poor solubility, enzymatic degradation, irregular distribution in the food matrix, and interactions between these natural additives and food components, food characteristics (pH, texture, etc.), and processing techniques. Optimization of the application conditions can help solve many of these drawbacks (Liu et al., 2021).
One of the strategies developed in recent years to reduce post-harvest losses of plantain in African is recipe development (Strano et al., 2022, Vida et al., 2024). These strategies, which include general recipes, site recipes, master recipes, and control recipes, were developed to minimised post-harvest losses especially in unripe plantain. However, only a limited number of recipes can be developed for unripe plantains, hence significant losses are still inevitable as most of the plantains become ripen under storage (Vida et al., 2024).
In order to decrease the post-harvest spoilage of plantain and enhance their safety, LAB based antifungal metabolites should be developed and effectively applied. Therefore, the present study was focused on evaluating the antifungal activities of LAB and their metabolites against post-harvest spoilage fungi of plantain with the aim of obtaining an alternative biopreservative approach for plantain.
Materials and methods
Materials
The materials used in this study included De Mann Rogosa Sharpe medium (MRS) (Himedia® M641), Potato Dextrose Agar (PDA) (Himedia® MH096), DNase agar (Thermo Scientific™ PO0128A), gelatinase agar (Merck DEV 1106850500), and Zymo Research Quick-DNA™ Miniprep Plus Kit (Zymo Research NC0933233).
Media preparation
MRS agar and PDA used for the isolation of LAB and moulds, respectively, were prepared as instructed by the manufacturer. The media were dissolved and sterilized at 121 oC for 15 min using the autoclave. Sterile media were cooled to about 45-50 oC before pouring into sterile Petri dishes.
Sample collection
A total of sixty samples of agadagidi and ogi were collected aseptically in sterile air tight containers with well fitted cover lids and transported to the laboratory for further analysis. Bunches of freshly harvested plantain with no observable sign of spoilage were collected from Ibadan metropolis, Nigeria, between February and April 2021, for this study, for this study. The fingers were sorted and cleaned under running tap water. These samples were surface sterilized with 1% sodium hypochlorite and stored in clean sterile containers at room temperature and relative humidity for 7-10 days until physical signs of mould spoilage appeared (Chuku, 2009).
Isolation and total count of LAB from agadagidi and ogi
Isolation of LAB from agadagidi and ogi was carried out using the pour plate method as described by Abdelkader et al. (2009). One ml from each of the inoculum dilutions was dispensed into sterile Petri dishes with duplicates prepared for each dilution, and approximately 10 ml of MRS agar, cooled to a temperature of about 45 oC, were added to each plate. The plates were allowed to set before been incubated anaerobically at 37 oC for 24 h in an inverted position. The colony count of each plate was done and the total load was calculated using the formula below:
Colony Forming Unit
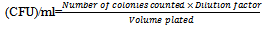
Distinct colonies were randomly selected and streaked on sterile MRS agar to achieve pure cultures. Gram staining and catalase test were carried out on the pure cultures to select presumptive LAB. The Gram-positive and catalase negative isolated were selected and kept on MRS agar slant and stored at 4 °C for short term storage and in MRS broth containing 20% glycerol stored at -4 oC for long term storage.
Fungal isolation
Colony Forming Unit
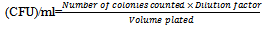
Distinct colonies were randomly selected and streaked on sterile MRS agar to achieve pure cultures. Gram staining and catalase test were carried out on the pure cultures to select presumptive LAB. The Gram-positive and catalase negative isolated were selected and kept on MRS agar slant and stored at 4 °C for short term storage and in MRS broth containing 20% glycerol stored at -4 oC for long term storage.
Fungal isolation
Isolates of fungi were obtained from the fingers of plantain with visible signs of fungal spoilage. Isolation was carried out using both the surface wash-off water and the plantain pulp. The surface wash-off water was prepared by washing whole plantain samples with 100 ml of sterile water. In addition, one g of the plantain pulp was weighed and pulverized in 9 ml of sterile water before being serially diluted. One ml of the inoculum was plated on sterile PDA in duplicate using the pour plate method. Fifty mg of chloramphenicol was incorporated per litre of the medium to inhibit bacterial growth. The plates were incubated at 28 oC for 5-7 days. Distinct colonies that appeared after the incubation period were picked and further purified by stabbing on sterile agar plates. This was done twice to obtain pure culture. The pure cultures were grown on fresh medium of PDA agar slant and incubated at 28±2 oC for 5-7 days. These isolates were preserved at 4 oC in the refrigerator for further examination (Al-Hindi et al., 2011).
Total fungal count
Appropriate dilutions taken from the decimal dilutions of sample homogenates were plated using PDA and incubated at 28±2 oC for 5-7 days. Distinct colonies were then counted and recorded in CFU/ml of plantain sample using the formula below (Sofo et al., 2020).

Fungal characterization
The method described by Okafor and Eni (2018) was employed for the microscopic examination of the characteristics of the fungal isolates. Identification of the fungal isolates was based on the cultural characteristics and morphology of the hyphae and spores as observed under the microscope.

The method described by Okafor and Eni (2018) was employed for the microscopic examination of the characteristics of the fungal isolates. Identification of the fungal isolates was based on the cultural characteristics and morphology of the hyphae and spores as observed under the microscope.
Screening of the LAB isolates for antifungal activity
-Preparation of Cell-Free Supernatants (CFS) from the bacterial isolates
The LAB isolates were grown in MRS broth at 37 oC for 24 h. The CFS of the bacterial isolates were obtained by removing the bacterial cells through centrifugation at 4,000 rpm for 30 min. The clear supernatant of each isolate was filtered through sterile Whatman filter of 0.22 µm pore size (Jeong et al., 2023).
-Standardization of fungal spores
The spores of the fungi were harvested by introducing 5 ml of sterile distilled water onto a three to seven days old fungal colony plate and homogenized using sterile inoculating loop. Standardization was done using spectrophotometer at wavelength of 630 nm with the reference Optical Density (OD) ranging between 0.09 and 0.56. Adjusted suspensions were enumerated by plating on PDA. The spores were dislodged and diluted to 106. Aliquot of 0.1 ml was spread-plated onto the PDA plates with the sterile swab stick. Subsequently, the plates were incubated at 28 oC for three to five days. Observable colonies were counted as soon as growth appeared on the plates (Caligiore-Gei and Valdez, 2015).
The LAB isolates were grown in MRS broth at 37 oC for 24 h. The CFS of the bacterial isolates were obtained by removing the bacterial cells through centrifugation at 4,000 rpm for 30 min. The clear supernatant of each isolate was filtered through sterile Whatman filter of 0.22 µm pore size (Jeong et al., 2023).
-Standardization of fungal spores
The spores of the fungi were harvested by introducing 5 ml of sterile distilled water onto a three to seven days old fungal colony plate and homogenized using sterile inoculating loop. Standardization was done using spectrophotometer at wavelength of 630 nm with the reference Optical Density (OD) ranging between 0.09 and 0.56. Adjusted suspensions were enumerated by plating on PDA. The spores were dislodged and diluted to 106. Aliquot of 0.1 ml was spread-plated onto the PDA plates with the sterile swab stick. Subsequently, the plates were incubated at 28 oC for three to five days. Observable colonies were counted as soon as growth appeared on the plates (Caligiore-Gei and Valdez, 2015).
-Initial screening for antifungal activities
The initial screening of the LAB isolates for their inhibitory potential against the isolated fungus was carried out using agar overlay method against the dominant fungus. The LAB were lawn streaked on MRS agar with the pH of the medium maintained at 6.8 and incubation temperature of 37 oC for 48 h under anaerobic condition. Fungal spores were standardized to 3×106 spore per ml according to the method outlined by Caligiore-Gei and Valdez (2015) and Petrikkou et al. (2001). Inhibition of the spore growth was determined and scored according to the method of Luz et al. (2017). The zones of inhibition’s diameters were measured and recorded in millimetre using a ruler. The score was assigned as follows: (−) indicated no inhibition, (+) represented an inhibition zone <7 mm, (++) denoted a zone between 7 mm and 14 mm, and (+++) corresponded to a zone ≥14 mm.
Determination of the inhibitory spectrum of the LAB using agar overlay method
The antifungal spectra of the selected LAB against the predominant moulds were evaluated using the agar overlay method. MRS agar was poured and allowed to harden in sterile petri dish. Then, the LAB isolates were lawn streaked on agar plates and incubated at 37 oC for 48 h under anaerobic condition. Lastly, the plates were overlaid with potato dextrose soft agar containing 3×106 spores/ml and incubated under aerobic condition at 28±2 oC for 48 h (Zebboudj et al., 2014).
Determination of the inhibitory spectrum of the CFS using agar well diffusion method
Wells of 5 ml diameter were punched into solidified PDA plates, already seeded with the standardized fungal spores, using a sterile stainless-steel borer. The wells were sealed with agar-agar at the base to avoid the escape of the CFS into the surrounding medium. Varying dilution ratios of CFS, 100 (1:0), 50 (1:1), and 25% (1:3), with sterile MRS broth as the diluent was used. The wells were filled with 0.1 ml of the CFL and allowed to diffuse into the agar for about 5 h at room temperature (pre-incubation period). Following this, the plates were incubated at 28±2 oC for 48 h. Lastly, the inhibition zone around the well were measured and graded according to Wang et al. (2012).
Phenotypic characterization of the LAB with antifungal activity
-Morphological and biochemical characterization of the isolates
The gram staining was carried out to determine the morphology of the LAB isolates. Later, spore formation and motility tests were carried out as described by Olutiola et al. (2000). Biochemical tests, including catalase, oxidase, citrate utilization, urease, and sugar fermentation, were carried out using standard protocols (Olutiola et al., 2000). The biochemical analysis results were used to identify the closest matching known bacterial genera, with taxonomic assignments based on Bergey's Manual of Systematic Bacteriology (Rose-Monde et al., 2013).
Phenotypic characterization of the LAB with antifungal activity
-Morphological and biochemical characterization of the isolates
The gram staining was carried out to determine the morphology of the LAB isolates. Later, spore formation and motility tests were carried out as described by Olutiola et al. (2000). Biochemical tests, including catalase, oxidase, citrate utilization, urease, and sugar fermentation, were carried out using standard protocols (Olutiola et al., 2000). The biochemical analysis results were used to identify the closest matching known bacterial genera, with taxonomic assignments based on Bergey's Manual of Systematic Bacteriology (Rose-Monde et al., 2013).
Safety assessment tests for the LAB isolates with antifungal activity
-DNase tests
This is a differential test that detects whether an organism can produce an exoenzyme called deoxyribonuclease (DNase), which hydrolyses DNA. Cultures of the test isolates were streaked on DNase agar plates and anaerobically incubated for 24 h. The plates were then flooded with 1% hydrogen chloride solution and observed for zone of clearance, which indicate DNase production (Cheesebrough, 2006).
This is a differential test that detects whether an organism can produce an exoenzyme called deoxyribonuclease (DNase), which hydrolyses DNA. Cultures of the test isolates were streaked on DNase agar plates and anaerobically incubated for 24 h. The plates were then flooded with 1% hydrogen chloride solution and observed for zone of clearance, which indicate DNase production (Cheesebrough, 2006).
-Gelatinase test
Smears of 24 h old cultures of test isolates were made on gelatine agar plates. The plates were incubated at 37 °C for 24 h before being flooded with acid mercury chloride and observed for zones of clearance around the bacterial colonies, which indicates a gelatin liquefying bacteria. However, non-liquefying bacteria produced no zone around its colony (Beshiru et al., 2018).
-Haemolysis test
Haemolysis test was carried out to provide information on what haemolytic enzyme is present in a bacterium by providing a culture enriched with red blood cells. Cultures that have been grown for 24 h were used for this test. The cultures were cross inoculated on blood agar plates and incubates at 30 oC for 24 h. Haemolytic bacteria are either α-(alpha), β (beta), or γ (gamma) haemolytic. Alpha (α) haemolysis is said to occur when red blood cells are partially destroyed, presenting a greenish-brownish zone around the inoculated area. The β haemolysis take place when the red blood cells are completely destroyed with clear zone around the inoculated area; while it is said to be γ haemolysis when there is no breakdown of the red blood cells (Erika et al., 2013).
Molecular characterization of the LAB with antifungal activity
-Bacterial DNA extraction and Polymerase Chain Reaction (PCR) amplification
Extraction of the LAB DNA was done using ZR BashingTM Lysis Tube and it was carried out according to the manufacturer’s instruction. This protocol was previously described by Onipede et al. (2020). The 16S rRNA gene from genomic DNA was amplified by PCR using bacteria universal primer pair 16SF: GTGCCAGCAGCCGCGCTAA and 16SR: AGACCCGGGAACGTATTCAC. The PCR amplification was carried out in a 10 µl reaction containing 1.0 µl of 10× PCR buffer, 1.0 µl of 25 mm MgCl2, 0.5 µl of 5 pMol of both forward primer and reverse primer, 1.0 µl of DMSO, 0.8µl of 2.5 mm DNTPs, 0.1 µl of Taq 5 µg/µl, 2.0 µl of 10 ng/µl DNA, and 3.1 µl of water was added in a PCR tube. PCR was carried out at an initial denaturation step at 94 oC for 5 min followed by 30 cycles at 94 oC for 30 s, 56 oC for 30 s, 72 oC for 45 s, and a final extension step at 72 oC for 7 min. The amplicon from the reaction was separated by electrophoresis on a 1.5% agarose gel.
-Haemolysis test
Haemolysis test was carried out to provide information on what haemolytic enzyme is present in a bacterium by providing a culture enriched with red blood cells. Cultures that have been grown for 24 h were used for this test. The cultures were cross inoculated on blood agar plates and incubates at 30 oC for 24 h. Haemolytic bacteria are either α-(alpha), β (beta), or γ (gamma) haemolytic. Alpha (α) haemolysis is said to occur when red blood cells are partially destroyed, presenting a greenish-brownish zone around the inoculated area. The β haemolysis take place when the red blood cells are completely destroyed with clear zone around the inoculated area; while it is said to be γ haemolysis when there is no breakdown of the red blood cells (Erika et al., 2013).
Molecular characterization of the LAB with antifungal activity
-Bacterial DNA extraction and Polymerase Chain Reaction (PCR) amplification
Extraction of the LAB DNA was done using ZR BashingTM Lysis Tube and it was carried out according to the manufacturer’s instruction. This protocol was previously described by Onipede et al. (2020). The 16S rRNA gene from genomic DNA was amplified by PCR using bacteria universal primer pair 16SF: GTGCCAGCAGCCGCGCTAA and 16SR: AGACCCGGGAACGTATTCAC. The PCR amplification was carried out in a 10 µl reaction containing 1.0 µl of 10× PCR buffer, 1.0 µl of 25 mm MgCl2, 0.5 µl of 5 pMol of both forward primer and reverse primer, 1.0 µl of DMSO, 0.8µl of 2.5 mm DNTPs, 0.1 µl of Taq 5 µg/µl, 2.0 µl of 10 ng/µl DNA, and 3.1 µl of water was added in a PCR tube. PCR was carried out at an initial denaturation step at 94 oC for 5 min followed by 30 cycles at 94 oC for 30 s, 56 oC for 30 s, 72 oC for 45 s, and a final extension step at 72 oC for 7 min. The amplicon from the reaction was separated by electrophoresis on a 1.5% agarose gel.
-PCR product purification
The process of purification started with adding 20 µl of absolute ethanol to the PCR product, followed by incubation at ambient temperature for 15 min. The mixture was spun at 10,000 rpm for 15 min; the supernatant decanted. The residue was also spun at 10,000 rpm for 15 min; the supernatant was decanted and 40 µl of 70% ethanol was added. Next, the product was air dried, and then about 10 µl of ultrapure water was added. The amplicon checked on a 1.5% agarose (Ma and Difazio, 2008).
-DNA sequencing and analysis
The genomic DNAs of the LAB were sequenced with 518F and 800R primers using ABI PRISM Big Dye Terminator cycle sequencer (Macrogen, USA). The comparison of the partial 16S rRNA gene sequence of the isolates to those available in the GenBank nucleotides sequence data-base was performed using BLAST. (http://www.nbci.nlm.nih.gov/Blast.cgi/) (Shehata et al., 2019).
-Evolutionary analysis by Maximum Likelihood method
The evolutionary history of the LAB was inferred based on the Maximum Likelihood using the Hasegawa-Kishino-Yano model (Hasegawa et al., 1985). The MEGA X software described by Kumar et al. (2018) was used for the analysis.
Statistical analysis
All quantitative data for the experiments were obtained in triplicates. Using IBM SPSS 2022, descriptive analysis based on one-way analysis of variance (ANOVA) at 95% confidence level (i.e., p≤0.05), was employed to compare the means and determine significant differences between treatments.
Results and discussion
Seventy-five presumptive LAB isolates, which were gram-positive and catalase-negative, were obtained from agadagidi and ogi. Fifty-one of the isolates were obtained from agadagidi while 24 were obtained from ogi. The highest total LAB count of 5.6 log10 CFU/ml was observed in ogi, while the lowest count was in agadagidi with a value of 3.7 log10 CFU/ml (Table 1). Sixteen fungal isolates were obtained from the plantain samples. Six of these were isolated from the pulp, while ten were isolated from the surface wash-off water. The total fungal count ranged from 2.89 log10 spore/g to 5.30 log10 spore/g (Table 1). The total LAB count obtained from agadagidi ranged from 3.7 log10 CFU/ml to 4.9 log10 CFU/ml. This is comparable to the report by Oriola et al. (2017), who recorded LAB in agadagidi. The higher load observed from the samples obtained from Masifa and Agurodo may be due to the long length of fermentation. Furthermore, the hygiene of the production environment where fermentation was carried out might also have influenced the observed LAB count. The LAB load obtained from ogi ranged from 4.9 log10 CFU/ml to 5.6 log10 CFU/ml. The processing and handling of ogi under unhygienic condition, where good manufacturing practices are lacking, might have contributed to the high bacterial load observed in samples.
The fungi were identified using features of the colony morphology and microscopic nature of the hyphal and spores. The morphological features, microscopic structure, and the organisms identified are presented in Table 2. The fungi were identified as A. flavus, Fusarium verticilliodes, Penicillium spp., Fusarium oxysporum, and Rhizopus sp. The highest occurring fungi was F. verticillioides (n=6; 37.5%), while the least occurring were A. flavus (n=1; 6.25%) and Rhizopus sp. (n=1; 6.25%) as presented in Figure 1. The high fungal load observed from the plantain samples indicated that possible post-harvest spoilage must have resulted from fungal activities. This agrees with earlier reports of Ajayi (2016) and Mogaji et al. (2021). These researchers recorded the prevalence of fungi and therefore considered fungi as spoilage organisms in plantain, attributing no significant role to them in the fermentation process during the production of agadagidi. The contributing factors to the rapid spoilage of plantains likely included lack of proper hygiene of the storage facilities as well as post-harvest exposure to contaminants from handlers, which might have influenced both the amount and the types of spoilage fungal species. The fungal isolates obtained and identified for this study included A. flavus, Fusarium verticillioides, F. oxysporum, Rhizopus sp., and Penicillium sp. Some of these fungi are known to be surface contaminants of many agricultural products that induce spoilage, while others were reportedly found in the environment where they induce storage spoilage of grains and are occasionally found in association with fruits. This result corresponds with the report of Ajayi (2016), who reported the presence of A. flavus, Fusarium sp, Rhizopus species, and Penicillium sp during a microbiological study of plantain. The presence of F. oxysporum, Rhizopus species and F. verticilliodes (moniliforme) in plantain was reported by Udoh et al. (2015) in the studies on fungi which cause the spoilage of edible fruits and vegetables. Similarly, the report by Temilade et al. (2020) implicated A. flavus as the predominant fungi in plantain flour samples.
The results obtained from screening the LAB isolates for antifungal activity against spoilage moulds using the agar overlay method, specifically the antifungal activity of isolate 5AG8 against the growth of F. oxysporum, are shown in Figure 2. The agar overlay method produced good activity and clear zone of inhibition after 48 h of incubation, and remained cleared for more than 14 days before being overcrowded by the growing mould. In contrast, the agar well diffusion technique used to assay the CFS activities, produced cleared zone of inhibition after 48 h of incubation (Figure 3), but became overcrowded with the fungi after five days. The initial screening results showed that 26 (34.67%) of the isolates have antifungal activity. The use of LAB as the bioprotective agent is one of the oldest and best approaches to food biopreservation with enormous potential characteristics such as low toxicity and high specificity (Luz et al., 2017). The antifungal activities of the LAB isolated from this study is similar to that reported by earlier researchers including Luz et al. (2017), Wang et al. (2012), Zebboudj et al. (2014). Both agar overlay and agar well method showed good results. However, better results were observed in the overlay method. L. plantarum has been reportedly identified to exhibit antifungal property similar to that of probiotic Lactobacillus strains (Arasu et al., 2013).
In this study, Penicillium sp. was the most susceptible to the LAB antifungal metabolites and A. flavus was the least inhibited by L. plantarum OP and L. fermentum 5AG8. Data related to the growth inhibition patterns of A. flavus, Penicillium sp., F. verticillioides, F. oxysporum, and Rhizopus sp., following exposure to varying concentrations of the culture filtrate supernatant, showed that L. fermentum 5AG8 exhibited the strongest antifungal potential against a wide range of moulds; with the highest inhibition produced being against F. oxysporum. This observation in the present study is comparable to that reported by Chen et al. (2021), who evaluated the antifungal activity of the CFS from two strains of LAB from kefir and reported that the CFS showed antifungal activity against Penicillium expansum. None of the LAB isolates showed significant antifungal activity against Rhizopus sp., even at all tested concentrations of the CFS, which contrasts with the clear dose-dependent antifungal response demonstrated by the CFS of the nine isolates against A. flavus, F. oxysporum, F. verticillioides, and Penicillium sp. The antifungal activity of the CFS was more pronounced against F. oxysporum followed by Penicillium sp., F. verticilliodes, and A. flavus. These results suggested that the isolates might have produced diffusible bioactive metabolites that are capable of exerting inhibitory effect on spoilage fungi. Mani-López et al. (2022) documented the activity of CFS from LAB against fungi. The CFS of LAB is known to produce strong inhibition of moulds on foods when applied directly or in packaged foods.
Inhibitory effect against spore germination and mycelial growth in agar well diffusion methods showed that inhibition was not well pronounced against mycelia growth. This showed that fungal spores were more susceptible to the bacterial metabolites. These observations are comparable to the report of Luz et al. (2017). Kivanc et al. (2014) also reported that LAB isolated from fermented guava juice were able to inhibit germination of spores and mycelial growth. Penicillium sp. spores were observed to be more susceptible to the activities of the LAB CFS, while A. flavus appeared the least susceptible to the treatment.
Statistical analysis
All quantitative data for the experiments were obtained in triplicates. Using IBM SPSS 2022, descriptive analysis based on one-way analysis of variance (ANOVA) at 95% confidence level (i.e., p≤0.05), was employed to compare the means and determine significant differences between treatments.
Results and discussion
Seventy-five presumptive LAB isolates, which were gram-positive and catalase-negative, were obtained from agadagidi and ogi. Fifty-one of the isolates were obtained from agadagidi while 24 were obtained from ogi. The highest total LAB count of 5.6 log10 CFU/ml was observed in ogi, while the lowest count was in agadagidi with a value of 3.7 log10 CFU/ml (Table 1). Sixteen fungal isolates were obtained from the plantain samples. Six of these were isolated from the pulp, while ten were isolated from the surface wash-off water. The total fungal count ranged from 2.89 log10 spore/g to 5.30 log10 spore/g (Table 1). The total LAB count obtained from agadagidi ranged from 3.7 log10 CFU/ml to 4.9 log10 CFU/ml. This is comparable to the report by Oriola et al. (2017), who recorded LAB in agadagidi. The higher load observed from the samples obtained from Masifa and Agurodo may be due to the long length of fermentation. Furthermore, the hygiene of the production environment where fermentation was carried out might also have influenced the observed LAB count. The LAB load obtained from ogi ranged from 4.9 log10 CFU/ml to 5.6 log10 CFU/ml. The processing and handling of ogi under unhygienic condition, where good manufacturing practices are lacking, might have contributed to the high bacterial load observed in samples.
The fungi were identified using features of the colony morphology and microscopic nature of the hyphal and spores. The morphological features, microscopic structure, and the organisms identified are presented in Table 2. The fungi were identified as A. flavus, Fusarium verticilliodes, Penicillium spp., Fusarium oxysporum, and Rhizopus sp. The highest occurring fungi was F. verticillioides (n=6; 37.5%), while the least occurring were A. flavus (n=1; 6.25%) and Rhizopus sp. (n=1; 6.25%) as presented in Figure 1. The high fungal load observed from the plantain samples indicated that possible post-harvest spoilage must have resulted from fungal activities. This agrees with earlier reports of Ajayi (2016) and Mogaji et al. (2021). These researchers recorded the prevalence of fungi and therefore considered fungi as spoilage organisms in plantain, attributing no significant role to them in the fermentation process during the production of agadagidi. The contributing factors to the rapid spoilage of plantains likely included lack of proper hygiene of the storage facilities as well as post-harvest exposure to contaminants from handlers, which might have influenced both the amount and the types of spoilage fungal species. The fungal isolates obtained and identified for this study included A. flavus, Fusarium verticillioides, F. oxysporum, Rhizopus sp., and Penicillium sp. Some of these fungi are known to be surface contaminants of many agricultural products that induce spoilage, while others were reportedly found in the environment where they induce storage spoilage of grains and are occasionally found in association with fruits. This result corresponds with the report of Ajayi (2016), who reported the presence of A. flavus, Fusarium sp, Rhizopus species, and Penicillium sp during a microbiological study of plantain. The presence of F. oxysporum, Rhizopus species and F. verticilliodes (moniliforme) in plantain was reported by Udoh et al. (2015) in the studies on fungi which cause the spoilage of edible fruits and vegetables. Similarly, the report by Temilade et al. (2020) implicated A. flavus as the predominant fungi in plantain flour samples.
The results obtained from screening the LAB isolates for antifungal activity against spoilage moulds using the agar overlay method, specifically the antifungal activity of isolate 5AG8 against the growth of F. oxysporum, are shown in Figure 2. The agar overlay method produced good activity and clear zone of inhibition after 48 h of incubation, and remained cleared for more than 14 days before being overcrowded by the growing mould. In contrast, the agar well diffusion technique used to assay the CFS activities, produced cleared zone of inhibition after 48 h of incubation (Figure 3), but became overcrowded with the fungi after five days. The initial screening results showed that 26 (34.67%) of the isolates have antifungal activity. The use of LAB as the bioprotective agent is one of the oldest and best approaches to food biopreservation with enormous potential characteristics such as low toxicity and high specificity (Luz et al., 2017). The antifungal activities of the LAB isolated from this study is similar to that reported by earlier researchers including Luz et al. (2017), Wang et al. (2012), Zebboudj et al. (2014). Both agar overlay and agar well method showed good results. However, better results were observed in the overlay method. L. plantarum has been reportedly identified to exhibit antifungal property similar to that of probiotic Lactobacillus strains (Arasu et al., 2013).
In this study, Penicillium sp. was the most susceptible to the LAB antifungal metabolites and A. flavus was the least inhibited by L. plantarum OP and L. fermentum 5AG8. Data related to the growth inhibition patterns of A. flavus, Penicillium sp., F. verticillioides, F. oxysporum, and Rhizopus sp., following exposure to varying concentrations of the culture filtrate supernatant, showed that L. fermentum 5AG8 exhibited the strongest antifungal potential against a wide range of moulds; with the highest inhibition produced being against F. oxysporum. This observation in the present study is comparable to that reported by Chen et al. (2021), who evaluated the antifungal activity of the CFS from two strains of LAB from kefir and reported that the CFS showed antifungal activity against Penicillium expansum. None of the LAB isolates showed significant antifungal activity against Rhizopus sp., even at all tested concentrations of the CFS, which contrasts with the clear dose-dependent antifungal response demonstrated by the CFS of the nine isolates against A. flavus, F. oxysporum, F. verticillioides, and Penicillium sp. The antifungal activity of the CFS was more pronounced against F. oxysporum followed by Penicillium sp., F. verticilliodes, and A. flavus. These results suggested that the isolates might have produced diffusible bioactive metabolites that are capable of exerting inhibitory effect on spoilage fungi. Mani-López et al. (2022) documented the activity of CFS from LAB against fungi. The CFS of LAB is known to produce strong inhibition of moulds on foods when applied directly or in packaged foods.
Inhibitory effect against spore germination and mycelial growth in agar well diffusion methods showed that inhibition was not well pronounced against mycelia growth. This showed that fungal spores were more susceptible to the bacterial metabolites. These observations are comparable to the report of Luz et al. (2017). Kivanc et al. (2014) also reported that LAB isolated from fermented guava juice were able to inhibit germination of spores and mycelial growth. Penicillium sp. spores were observed to be more susceptible to the activities of the LAB CFS, while A. flavus appeared the least susceptible to the treatment.
Table 1: Total counts of Lactic Acid Bacteria (LAB) and fungi in ogi and agadagidi
| Organism | Sample code | Sampling location | Total count log10 CFU/ml |
| LAB from agadagidi and ogi | AG | Bodija | 3.70±0.24 |
| 2AG | Masifa | 4.90±0.17 | |
| 3AG | Masifa | 4.50±0.37 | |
| 4AG | Agurodo | 4.65±0.27 | |
| 5AG | Agurodo | 3.87±0.15 | |
| 1OG | Orogun | 5.60±0.23 | |
| 2OG | Bodija | 4.90±0.27 | |
| Count log10 spore/g | |||
| Fungi from plantain | CONT1 | Orogun | 4.60±0.11 |
| CONT2 | Bodija | 4.30±0.09 | |
| AFW1 | UI | 5.30±0.10 | |
| AFW2 | Bodija | 2.89±0.07 |
Mean±Standard Deviation (SD) errors at 95% confidence interval (p≤0.05)
AFW1, AFW2, CONT1, CONT2=codes for samples of plantain; AG=code for samples of agadagidi; OG=code for samples of ogi
Table 2: Morphological characteristics and probable identities of the fungal plantain
AFW1, AFW2, CONT1, CONT2=codes for samples of plantain; AG=code for samples of agadagidi; OG=code for samples of ogi
Table 2: Morphological characteristics and probable identities of the fungal plantain
| Colonial characteristics | Microscopic morphology | Probable organism |
| white-pink flat, cotton-like colonies, which developed a purple colour on the reverse side at old age. | The mycelia were hyaline and septate. The phialides were cylindrical bearing the sickle shaped conidia. The microconidia were in chain, clustered and formed from the monophailides. | Fusarium verticilliodes |
| Creamy small colony, which developed a lime green colour and a creamy reverse as the colonies age. | The conidiophores were straight and beared the phialides at their tips. Hyphae were septate and hyaline. The conidial head was radiate, while the conidia appeared smooth and globose. | Aspergillus flavus |
| A deep green colony with a white-cream ring around the central green mass. The reverse changed black as the colony grew older. | The conidiophore was highly branched with phialides,that proliferated into long, broad, thick, smooth-walled structures exhibiting a brownish colouration. | Penicillium species |
| The colony initially developed as a white cottony mycelium. As it aged, a pink center surrounded by white ring with dark purple on the reverse side developed. | Microconidia were oval and the macroconidia were also oval but had septal. | Fusarium oxysporium |
| Grey-white with black pigment at the top. Colony grew fast and covered plate within a short time on incubation. The reverse side of the plate was initially white but turned black as it aged. | The hyphae were non septate. At the tips of the sporangiosphores were sporangiospores. At the base of the sporangiosphores were rhizoids, which linked one to the other. |
| Rhizopus species |


Figure 2: Antifungal activity of Lactobacillus fermentum 5AG8 against the growth of Fusarium oxysporum using agar overlay method

The mean antifungal activities of the LAB isolates against the spoilage fungi are presented in Table 3. Nine isolates, which represent approximately 12% of the total number of the isolated LAB, were selected based on their high activity against the spoilage organisms with zones of inhibition of ≥14 mm. A. flavus and F. verticilloides were resistant to seven of the nine LAB isolates. F. oxysporum was resistant to five isolates, while Penicillium sp was resistant to four. However, Rhizopus sp was resistant to all the LAB isolates. One of the LAB isolates (5AG8) had activity against three (A. flavus, F. oxysporum, and Penicillium sp.) out of the five spoilage fungi. The evaluation of the antifungal activities of the CFS of LAB isolates against the spores and mycelial growth of the spoilage fungi is represented on Table 4. All the spoilage fungi were susceptible to the CFS of one or more of the LAB except Rhizopus sp., which was resistant to the CFS of all the LAB isolates.
Table 3: Inhibition (mm) of the spoilage fungi by the Lactic Acid Bacteria (LAB) isolates
| LAB isolates | Spoilage fungi | ||||
| Aspergillus flavus | Fusarium oxysporum | Fusarium verticilloides | Rhizopus sp | Penicillium sp | |
| OC | 0 a | 14.5±0.28 b | 0 a | 0 | 0 a |
| OG | 0 a | 15.0±0.37 c | 0 a | 0 | 0 a |
| OP | 14.0±0.27 b | 0 a | 0 a | 0 | 15.5±0.47 c |
| AG1 | 0 a | 0 a | 14.5±0.22 b | 0 | 17.5±0.57 e |
| AG20 | 0 a | 0 a | 0 a | 0 | 0 a |
| AG24 | 0 a | 21.0±0.66 d | 0 a | 0 | 0 a |
| 3AG1 | 0 a | 0 a | 14.5±0.26 b | 0 | 14.5±0.65 b |
| 3AG2 | 0 a | 0 a | 0 a | 0 | 17.0±0.34 d |
| 5AG8 | 15.0±0.45 c | 23.5±0.42 e | 0 a | 0 | 23.0±0.78 f |
0: no inhibition; Mean±Standard Deviation (SD) errors at 95% confidence interval (p≤0.05); Different superscripts attached to the mean in the columns represent significant difference (p≤0.05)
AG1, AG20, AG24, 3AG1, 3AG2, 5AG8= codes assigned to isolates form agadagidi; OC, OP, OP= codes assigned to isolates fron ogi
Table 4: Inhibition of spores and mycelial growth of the spoilage fungi by the Cell-Free Supernatants (CFS) of Lactic Acid Bacteria (LAB) isolates
AG1, AG20, AG24, 3AG1, 3AG2, 5AG8= codes assigned to isolates form agadagidi; OC, OP, OP= codes assigned to isolates fron ogi
Table 4: Inhibition of spores and mycelial growth of the spoilage fungi by the Cell-Free Supernatants (CFS) of Lactic Acid Bacteria (LAB) isolates
| LAB isolates | Spoilage fungal spores and mycelial growth | |||||||||
| Aspergillus flavus | Fusarium oxysporum | Fusarium verticilloides | Rhizopus sp | Penicillium sp | ||||||
| Spores | Mycelial growth | Spores | Mycelial growth | Spores | Mycelial growth | Spores | Mycelial growth | Spores | Mycelial growth | |
| OC | - | - | - | ++ | - | - | - | - | - | - |
| OG | - | - | - | ++ | - | - | - | - | - | - |
| OP | ++ | - | - | - | - | - | - | - | +++ | ++ |
| AG1 | - | - | - | - | ++ | ++ | - | - | +++ | ++ |
| AG20 | - | - | ++ | - | - | ++ | - | - | - | +++ |
| AG24 | - | - | +++ | ++ | - | - | - | - | - | - |
| 3AG1 | - | - | - | - | ++ | - | - | - | +++ | + |
| 3AG2 | - | - | - | - | - | - | - | - | +++ | ++ |
| 5AG8 | +++ | ++ | +++ | ++ | - | - | - | - | +++ | ++ |
-: no inhibition; +: inhibition <7mm; ++: inhibition ≥7mm<14mm; +++: inhibition ≥14mm
AG1, AG20, AG24, 3AG1, 3AG2, 5AG8= codes assigned to isolates form agadagidi; OC, OP, OP= codes assigned to isolates fron ogi
AG1, AG20, AG24, 3AG1, 3AG2, 5AG8= codes assigned to isolates form agadagidi; OC, OP, OP= codes assigned to isolates fron ogi
The nine selected LAB isolates were further characterized to identify them. All the nine isolates were rod shaped, non-sporing, non-motile, and negative for catalase, oxidase, citrate utilization, and urease tests, making them probable LAB. The phenotypic characteristics of LAB isolates with antifungal activity, with regards to their morphology and sugar fermentation pattern, are presented in Table 5. The sugar fermentation array of the bacterial isolates showed that the LAB isolates fermented glucose, sucrose, and maltose. With the exception of AG1 and 3AG1, all the isolates fermented mannitol. Galactose was utilized by all the isolates except OG, AG24, and 3AG1; while OC, OG, and 3AG1 could not use lactose. The probable identity of the bacteria was based on Bergey’s manual of systematic bacteriology, which indicated all the suspected LAB as Lactobacillus species. Isolation of LAB from ogi was considered appropriate since ogi fermentation was primarily carried out by LAB. The identified bacterial isolates, using 16S rRNA sequencing, included Lactobacillus plantarum AG1 and Lactobacillus fermentum 5AG8 isolated from agadagidi, while L. plantarum OP was isolated from ogi. Isolation of L. fermentum and L. plantarum were identified in agadagidi by Oriola et al. (2017), with a high chance of occurrence after 24 and 48 h of fermentation. Furthermore, the results of the safety assessment tests revealed that all isolates are safe for use in food matrices; as all tested LAB strains showed γ-haemolysis with no breakdown of red blood cells, indicating their non-haemolytic characteristic. Similarly, negative results were obtained from the DNase and gelatinase test. These results were consistent with earlier reports of the Generally Regarded As Safe (GRAS) status of LAB and the safety of their application in food matrices (Amenu and Bacha, 2023; Oyewole et al 2018). Lahiri et al. (2021) opined that LAB are known to be the safest group of organisms used in the food industry. One of the mechanisms used by LAB for their actions against pathogenic microorganisms is the production of bacteriocin, a metabolite with the ability to decrease the pH of the food matrices, inhibiting the growth of a wide range of food spoilage microorganisms (Da Costa et al., 2019; Lahiri et al., 2021).
Based on the antifungal activity among the nine LAB isolates, OP, AG1, and 5AG8, which had marked inhibitory activity against two or more spoilage fungi, were identified using 16S rRNA sequencing and compared with sequences obtainable in GenBank nucleotide database. Isolate OP and AG1 were identified as L. plantarum, while 5AG8 was identified as L. fermentum. The maximum likelihood phylogenetic tree (Figure 4) shows that the two L. plantarum strains from this study are closely related, having high homology of 90%; while the L. fermentum 5AG8 shared 84% homology with the two L. plantarum strains and a reference strain from the GenBank.
Based on the antifungal activity among the nine LAB isolates, OP, AG1, and 5AG8, which had marked inhibitory activity against two or more spoilage fungi, were identified using 16S rRNA sequencing and compared with sequences obtainable in GenBank nucleotide database. Isolate OP and AG1 were identified as L. plantarum, while 5AG8 was identified as L. fermentum. The maximum likelihood phylogenetic tree (Figure 4) shows that the two L. plantarum strains from this study are closely related, having high homology of 90%; while the L. fermentum 5AG8 shared 84% homology with the two L. plantarum strains and a reference strain from the GenBank.
Table 5: Phenotypic characteristics of Lactic Acid Bacteria (LAB) Isolates with antifungal activity
| Isolate code | Gram reaction/ Morphology |
Sugar fermentation pattern | Probable Organism | ||||||
| Manitol | Glucose | Sucrose | Galactose | Lactose | Maltose | Fructose | |||
| OC | + SR | + ng | + ng | + ng | + ng | - | + ng | + ng | Lactobacillus sp |
| OG | + SR | + ng | + ng | + ng | - | - | + ng | + ng | Lactobacillus sp |
| OP | + LR | + ng | + ng | + ng | + ng | + ng | + ng | + ng | Lactobacillus sp |
| AG1 | + SR | - | + ng | + ng | + ng | + ng | + ng | + ng | Lactobacillus sp |
| AG20 | + LR | + ng | + ng | + ng | + ng | + ng | + ng | + ng | Lactobacillus sp |
| AG24 | + SR | + ng | + ng | + ng | - | + ng | + ng | - | Lactobacillus sp |
| 3AG1 | + LR | - | + ng | + ng | - | - | + ng | + ng | Lactobacillus sp |
| 3AG2 | + LR | + ng | + ng | + ng | + ng | + ng | + ng | + ng | Lactobacillus sp |
| 5AG8 | + LR | + ng | + ng | + ng | + ng | + ng | + ng | + ng | |
| Lactobacillus sp |
SR=Short Rods; LR=Long Rods
+=Positive; -=Negative; ng=No gas production
AG1, AG20, AG24, 3AG1, 3AG2, 5AG8= codes assigned to isolates form agadagidi; OC, OP, OP= codes assigned to isolates fron ogi
+=Positive; -=Negative; ng=No gas production
AG1, AG20, AG24, 3AG1, 3AG2, 5AG8= codes assigned to isolates form agadagidi; OC, OP, OP= codes assigned to isolates fron ogi

Figure 4: Evolutionary analysis constructed using the maximum likelihood technique
Conclusion
The isolated LAB demonstrated optimum antifungal activity against the spoilage fungi isolated from plantain. This study revealed that L. plantarum AG1, L. plantarum OP, and L. fermentum 5AG8, sourced from agadagidi and ogi, have the potential to inhibit spoilage fungi in plantain. LAB from other local fermented foods could also be explored for their potentials as biopreservatives in products prone to post-harvest fungal spoilage, such as fruits and vegetables. Profiling the metabolites in the CFS of these LAB strains could also help to elucidate the active compounds of the metabolites. Safety analysis results indicated that all the isolates are non-haemolytic and may be considered safe for use in food matrices, supporting their application as biopreservative in plantain.
Author contributions
G.O.O. and A.I.S. conceptualize and designed this study; E.K.O. conducted the experimental work, analyzed the data and wrote the first draft of the manuscript; the correction of the manuscript was done by G.O.O. and final draft and approval of the version to be submitted was done by A.I.S. All authors read and approved the final manuscript.
Acknowledgments
The authors acknowledge the technical staff of the Department of Microbiology, University of Ibadan, Nigeria for their support during the bench work of this study.
Conflicts of interest
The authors declare that there is no conflict of interest.
Funding
This research received no specific grant from any funding agency in the public, commercial, or non-profit sectors.
Ethical consideration
Not applicable.
The isolated LAB demonstrated optimum antifungal activity against the spoilage fungi isolated from plantain. This study revealed that L. plantarum AG1, L. plantarum OP, and L. fermentum 5AG8, sourced from agadagidi and ogi, have the potential to inhibit spoilage fungi in plantain. LAB from other local fermented foods could also be explored for their potentials as biopreservatives in products prone to post-harvest fungal spoilage, such as fruits and vegetables. Profiling the metabolites in the CFS of these LAB strains could also help to elucidate the active compounds of the metabolites. Safety analysis results indicated that all the isolates are non-haemolytic and may be considered safe for use in food matrices, supporting their application as biopreservative in plantain.
Author contributions
G.O.O. and A.I.S. conceptualize and designed this study; E.K.O. conducted the experimental work, analyzed the data and wrote the first draft of the manuscript; the correction of the manuscript was done by G.O.O. and final draft and approval of the version to be submitted was done by A.I.S. All authors read and approved the final manuscript.
Acknowledgments
The authors acknowledge the technical staff of the Department of Microbiology, University of Ibadan, Nigeria for their support during the bench work of this study.
Conflicts of interest
The authors declare that there is no conflict of interest.
Funding
This research received no specific grant from any funding agency in the public, commercial, or non-profit sectors.
Ethical consideration
Not applicable.
References
Abdelkader M., Chihib N.E., Abdelkader D.B., Naima N.A., Jean P.H. (2009). Antibacterial activity of some lactic acid bacteria isolated from an Algerian dairy product. Journal of Environmental and Public Health. 1-6. [DOI: 10.1155/2009/678495]
Abdu M.M., Rashid S., Beatrice K. (2024). Determination of aflatoxin level in stored maize flour and risk of exposure from consumption of aflatoxin contaminated meal in boarding school. International Journal of Microbiology and Biotechnology. 9: 7-14. [DOI: 10.11648/ijmb.20240901.12]
Abiodun-Solanke A.O., Falade K.O. (2010). A review of the uses and methods of processing banana and plantain (Musa spp.) into storable food products. Journal of Agricultural Research and Development. 9. [DOI: 10.4314/jard.v9i2.66815]
Ajayi A.O. (2016). Microbiological quality of plantain (musa paradisiacal). Nigerian Journal of Microbiology. 30: 3611-3618.
Al-Hindi R.R., Al-Najada A.R., Mohamed S.A. (2011). Isolation and identification of some fruit spoilage fungi: screening of plant cell wall degrading enzymes. African Journal of Microbiology Research. 5: 443-448. [DOI: 10.5897/AJMR10.896]
Amenu D., Bacha K. (2023). Probiotic potential and safety analysis of lactic acid bacteria isolated from Ethiopian traditional fermented foods and beverages. Annals of Microbiology. 73: 37. [DOI: 10.1186/s13213-023-01740-9]
Arasu M.V., Jung M.-W., Ilavenil S., Jane M., Kim D.-H., Lee K.-D., Park H.-S., Hur T.-Y., Choi G.-J., Lim Y.-C., Al-Dhabi N.A., Choi K.-C. (2013). Isolation and characterization of antifungal compound from Lactobacillus plantarum KKC-10 from forage silage with potential beneficial properties. Journal of Applied Microbiology. 115: 1172-1185. [DOI: 10.1111/jam.12319]
Begley M., Hill C., Gahan C.G.M. (2006). Bile salt hydrolase activity in probiotics. Applied and Environmental Microbiology. 72: 1729-1738. [DOI: 10.1128/AEM.72.3.1729-1738.2006]
Beshiru A., Okareh O.T., Chigor V.N., Igbinosa E.O. (2018). Assessment of water quality of rivers that serves as water sources for drinking and domestic functions in rural and urban communities in Edo North, Nigeria. Environmental Monitoring and Assessment. 190:387. [DOI: 10.1007/s10661-018-6771-7]
Bin Masalam M.S., Bahieldin A., Alharbi M.G., Al-Masaudi S., Al-Jaouni S.K., Harakeh S.M., Al-Hindi R.R. (2018). Isolation, molecular characterization and probiotic potential of lactic acid bacteria in Saudi raw and fermented milk. Evidence-Based Complementary and Alternative Medicine. 2018: 7970463. [DOI: 10.1155/2018/7970463]
Caligiore-Gei P.F., Valdez J.G. (2015). Adjustment of a rapid method for the quantification of Fusarium spp. spore suspensions in plant pathology. Revista Argentina de Microbiología. 47: 152-154. [DOI: 10.1016/j.ram.2015.03.002]
Chavan P., Lata K., Kaur T., Rezek Jambrak A., Sharma S., Roy S., Sinhmar A., Thory R., Pal Singh G., Aayush K., Rout A. (2023). Recent advances in the preservation of postharvest fruits using edible films and coatings: a comprehensive review. Food Chemistry. 418: 135916. [DOI: 10.1016/j.foodchem. 2023.135916]
Cheesebrough M. (2006). District laboratory practice in tropical countries. 2nd edition. Cambridge University Press, Cambridge, UK. pp: 1-434. [DOI: 10.1017/CBO9780511543470]
Chen H., Ju H., Wang Y., Du G., Yan X., Cui Y., Yuan Y., Yue T. (2021). Antifungal activity and mode of action of lactic acid bacteria isolated from kefir against Penicillium expansum. Food Control. 130: 108274. [DOI: 10.1016/j.foodcont.2021.108274]
Chuku E.C. (2009). Fungi responsible for the spoilage of plantain (Musa paradisiaca) at different ripening stage. Acta Agronomica Nigeriana. 9: 35-40.
Crowley S., Mahony J., van Sinderen D. (2013). Current perspectives on antifungal lactic acid bacteria as natural bio-preservatives. Trends in Food Science and Technology. 33: 93-109. [DOI: 10.1016/j.tifs.2013.07.004]
Da Costa R.J., Voloski F.L.S., Mondadori R.G., Duval E.H., Fiorentini Â.M. (2019). Preservation of meat products with bacteriocins produced by lactic acid bacteria isolated from meat. Journal of Food Quality. 4726510. [DOI: 10.1155/2019/4726510]
Eni A.O., Oluwawemitan I.A., Solomon O.U. (2010). Microbial quality of fruits and vegetables sold in Sango Ota, Ogun State, Nigeria. African Journal of Food Science. 4: 291-296.
Erika M.T., Andrea K.B., Tamás F., Balázs V., Rita S., Károly M. (2013). Practical microbiology. Eötvös Loránd University, Budapest, Hungary. pp: 1-222. URL: https://ttk.elte.hu/dstore/document/893/book.pdf.
Hasegawa M., Kishino H., Yano T. (1985). Dating the human-ape split by a molecular clock of mitochondrial DNA. Journal of Molecular Evolution. 22: 160-174. [DOI: 10.1007/BF02101694]
Honfo F.G., Kayodé A.P.P., Coulibaly O., Tenkouano A. (2007). Relative contribution of banana and plantain products to the nutritional requirements for iron, zinc and vitamin A of infants and mothers in Cameroon. Fruits. 62: 267-277. [DOI: 10.1051/fruits:2007023]
Huan Y., Kong Q., Mou H., Yi H. (2020). Antimicrobial peptides: classification, design, application and research progress in multiple fields. Frontier in Microbiology. 11: 582779. [DOI: 10.3389/fmicb.2020.582779]
International Institute of Tropical Agriculture (IITA). (2000). Improving plantain and banana based. International Institute of Tropical Agriculture Annual Report. 13-14. URL: https:// www.iita.org/wp-content/uploads/2016/04/Annual-Report-2000-full-version.pdf.
Jeong S.-G., Kim H.M., Lee M., Yang J.E., Park H.W. (2023). Use of vegetable waste as a culture medium ingredient improves the antimicrobial and immunomodulatory activities of Lantiplantibacillus plantarum Wikim0125 isolated form kimchi. Journal of Microbiology and Biotechnology 33: 75-82. [DOI: 10.4014/jmb.2210.10049]
Kivanc M., Kivanc S.A., Pektas S. (2014). Screening of lactic acid bacteria for antifungal activity against fungi. Journal of Food Processing and Technology. 5: 1-4. [DOI: 10.4172/2157-7110.1000310]
Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution. 35: 1547-1549. [DOI: 10.1093/molbev/msy096]
Lahiri D., Nag M., Sarkar T., Ray R.R., Shariati M.A., Rebezov M., Bangar S.P., Lorenzo J.M., Domínguez R. (2021). Lactic Acid Bacteria (LAB): autochthonous and probiotic microbes for meat preservation and fortification. Foods. 11: 2792. [DOI: 10.3390/foods11182792]
Liu J., Huang R., Song Q., Xiong H., Ma J., Xia R., Qiao J. (2021). Combinational antibacterial activity of nisin and 3-phenyllactic acid and their co-production by engineered lactococcus lactis. Frontiers in Bioengineering and Biotechnology. 9: 612105. [DOI: 10.3389/fbioe.2021.612105]
Luz C., Saladino F., Lusiano F.B., Mañes J., Meca G. (2017). Invitro antifungal activity of bioactive peptides produced by Lactobacillus plantarum against Aspergillus parasiticus and Penicillium expansum. LWT - Food Science and Technology. 81: 128-135. [DOI: 10.1016/j.lwt.2017.03.053]
Ma H., Difazio S. (2008). An efficient method for purification of PCR products for sequencing. Biotechniques. 44: 921-923. [DOI: 10.2144/000112809]
Mafe A.N., Edo G.I., Makia R.S., Joshua O.A., Akpoghelie P.O., Gaaz T.S., Jikah A.N., Yousif E., Isoje E.F., Igbuku U.A., Ahmed D.S., Essaghah A.E.A., et al. (2024). A review on food spoilage mechanisms, food borne diseases and commercial aspects of food preservation and processing. Food Chemistry Advances. 5: 100852. [DOI: 10.1016/j.focha.2024.100852]
Mani-López E., Arrioja-Bretón D., López-Malo A. (2022). The impacts of antimicrobial and antifungal activity of cell-free supernatants from lactic acid bacteria in vitro and foods. Comprehensive Reviews in Food Science and Food Safety. 21: 604-641. [DOI: 10.1111/1541-4337.12872]
Miedes E., Lorences E.P. (2004). Apple (Malus domestica) and tomato (Lycopersicum esculentum) fruits cell wall hemicelluloses and xyloglucan degradation during Penicillium expansum infection. Journal of Agriculture and Food Chemistry. 52: 7957-7963. [DOI: 10.1021/jf048890f]
Mogaji K.J., Arotupin D.J., Mogaji O., Arogunjo A.O., Ajayi-Moses O.B., Akinwunmi I.M., Gabriel P.O., Orekoya E.S., Olajesu O.L., Adeleye H.J., Aribisala J.O. (2021). Inherent microorganisms affects the quality of a Nieria fermented beverage “Agadagidi” during production. Asian Food Science Journal. 20: 6-17. [DOI: 10.9734/afsj/2021/v20i730316]
Ofosu D.O., Before I., Martinson F., Frimpong G.K., Asare I.K., Darfour B. (2023). Pre-harvest and post-harvest practices along the plantain (Musa spp. AAB) fruit value chain in Ghana that predispose them to ripening. Ghana Journal of Agricultural Science. 58: 75–82.
Okafor S.E., Eni A.O. (2018). Microbial quality and the occurrence of aflatoxins in plantain/yam and wheat flours in Ado-Odo Ota. Earth and Environmental Science. 210: 1-22. [DOI: 10.1088/1755-1315/210/1/012017]
Olutiola P.O., Famurewa, O., Suntay, H.G. (1991). An introduction to general microbiology: a practical approach. Hygiene Institut der Universitat Heidelberg. Heidelberg. URL: https://www.scirp.org/reference/referencespapers?referenceid=561804.
Onipede G., Aremu B., Sanni A., Babalola O. (2020). Molecular study of the phytase gene in lactic acid bacteria isolated from Ogi and Kunun-zaki, African fermented cereal gruel and beverage. Applied Food Biotechnology. 7: 49-60. [DOI: 10.22037/afb.v7i1.25909]
Oriola O., Boboye B., Adetuyi O. (2017). Microorganisms associated with the production of a Nigerian fermented beverage, ‘Agadagidi’. Microbiology Research Journal International. 20: 1-9. [DOI: 10.9734/MRJI/2017/32654]
Oyewole O.F., Maria C.O., Tope P.S., Funmi O.O. (2018). In vitro study of potential probiotic lactic acid bacteria isolated from the gut of chickens in Abeokuta, Nigeria. Alexandria Journal of Veterinary Science. 58: 73-84. [DOI: 10.5455/ajvs.290499]
Petrikkou E., Rodrı́guez-Tudela J.L., Cuenca-Estrella M., Gomez A., Molleja A., Mellado E. (2001). Inoculum standardization for antifungal susceptibility testing of filamentous fungi pathogenic for humans. Journal of Clinical Microbiology. 39: 1345-1347. [DOI: 10.1128/JCM.39.4.1345-1347.2001]
Ramudingana P., Mamphogoro T.P., Kamutando C.N., Maboko M.M., Modika K.Y., Moloto K.W., Thantsha M.S. (2024). Antagonistic potential of endophytic fungal isolates of tomato (Solanum lycopersicum L.) fruits against post-harvest disease-causing pathogens of tomatoes: An in vitro investigation. Fungal Biology. 128: 1847-1858. [DOI: 10.1016/j.funbio.2024.05.006]
Rios Covian D., Ruas-Madiedo P., Margolles A., Gueimonde M., De Los Reyes- Gavilán C.G., Salazar N. (2016). Intestinal short chain fatty acids and their link with diet and human health. Frontiers in Microbiology. 7: 185. [DOI: 10.3389/fmicb. 2016.00185]
Robinson J.C., Anderson T., Eckstein K. (1992). The influence of functional leaf removal at flower emergence on components of yield and photosynthetic compensation in banana. Journal of Horticultural Science. 67: 403-410. [DOI: 10.1080/00221589. 1992.11516265]
Rose-Monde M., Séverin K.K.A., Lessoy Z., Edwige A., Koffi Y., Sébastien N.L. (2013). Microbiological safety and quality assessment of a very appreciate traditional ready to eat plantain food, sold in retails markets. African Journal of Microbiology Research. 7: 4123-4129.
Rouse S., Harnett D., Vaughan A., Van Sinderen D. (2008). Lactic acid bacteria with potential to eliminate fungal spoilage in foods. Journal of Applied Microbiology. 104: 915-923. [DOI: 10.1111/j.1365-2672.2007.03619.x]
Salas M.L., Mounier J., Valence F., Coton M., Thierry A., Coton E. (2017). Antifungal microbial agents for food biopreservation—a review. Microorganisms. 5: 37. [DOI: 10.3390/ microorganisms5030037]
Shehata M.G., Badr A.N., El Sohaimy S.A., Asker D., Awad T.S. (2019). Characterization of antifungal metabolites produced by novel lactic acid bacterium and their potential application as food biopreservatives. Annals of Agricultural Sciences. 64: 71-78. [DOI: 10.1016/j.aoas.2019.05.002]
Silva C.C.G., Silva S.P.M., Ribeiro S.C. (2018). Application of bacteriocins and protective cultures in dairy food preservation. Frontier in Microbiology. 9: 594. [DOI: 10.3389/fmicb.2018.00594]
Sofo A., Elshafie H.S., Camele I. (2020). Structural and functional organization of the root system: a comparative study on five plant species. Plants. 9: 1338. [DOI: 10.3390/plants9101338]
Strano M.C., Altieri G., Allergra M., Di Renzo G.C., Paterna G., Matera A., Genovese F. (2022). Postharvest technologies of fresh citrus fruit: advances and recent developments for loss reduction during handling and storage. Horticulturae. 8: 612. [DOI: 10.3390/horticulturae8070612]
Temilade O.P., Olubukola O.J., Akinkunmi W.A. Rukayat H.O. (2020). Antibacterial sensitivity profile of Lactobacillus sp and characterization of aflatoxin-producing fungi isolated from yam and plantain flour. The International Journal of Biotechnology. 9: 14-23. [DOI: 10.18488/journal.57.2020.91.14.23]
Udoh I.P., Eleazar C.I., Ogeneh B.O., Ohanu M.E. (2015). Studies on fungi responsible for the spoilage/deterioration of some edible fruits and vegetables. Advances in Microbiology. 5: 285-290. [DOI: 10.4236/aim.2015.54027]
Vida C., Evelyn C.I., Akrofi-Ansah H., Edwina A., Beatrice Q. (2024). Food processing: reducing postharvest losses of plantain through alternative recipe development. Acta Scientific Nutritional Health. 8: 60-66.
Wang H., Yan Y., Wang J., Zhang H., Qi W. (2012). Production and characterization of antifungal compounds produced by Lactobacillus plantarum IMAU10014. PLOS ONE. 7: e29452. [DOI: 10.1371/journal.pone.0029452]
Zanotti I., Turroni F., Piemontese A., Mancabelli L., Milani C., Viappiani A., Prevedini G., Sanchez B., Margolles A., Elviri L., Franco B., Van Sinderen D., et al. (2015). Evidence for cholesterol lowering activity by Bifidobacterium bifidum PRL2010 through gut microbiota modulation. Applied Microbiology and Biotechnology. 99: 6813-6829. [DOI: 10.1007/s00253-015-6564-7]
Zapaśnik A., Sokołowska B., Bryła M. (2022). Role of lactic acid bacteria in food preservation and safety. Foods. 11: 1283. [DOI: 10.3390/foods11091283]
Zebboudj N., Yezli W., Hamini-Kadar N., Kihal M., Henni J.E. (2014). Antifungal activity of lactic acid bacteria against Fusarium oxysporum f. sp. albedinis isolated from diseased date palm in South Algeria. International Journal of Biosciences. 5: 99-106. [DOI: 10.22364/eeb.18.02]
Abdu M.M., Rashid S., Beatrice K. (2024). Determination of aflatoxin level in stored maize flour and risk of exposure from consumption of aflatoxin contaminated meal in boarding school. International Journal of Microbiology and Biotechnology. 9: 7-14. [DOI: 10.11648/ijmb.20240901.12]
Abiodun-Solanke A.O., Falade K.O. (2010). A review of the uses and methods of processing banana and plantain (Musa spp.) into storable food products. Journal of Agricultural Research and Development. 9. [DOI: 10.4314/jard.v9i2.66815]
Ajayi A.O. (2016). Microbiological quality of plantain (musa paradisiacal). Nigerian Journal of Microbiology. 30: 3611-3618.
Al-Hindi R.R., Al-Najada A.R., Mohamed S.A. (2011). Isolation and identification of some fruit spoilage fungi: screening of plant cell wall degrading enzymes. African Journal of Microbiology Research. 5: 443-448. [DOI: 10.5897/AJMR10.896]
Amenu D., Bacha K. (2023). Probiotic potential and safety analysis of lactic acid bacteria isolated from Ethiopian traditional fermented foods and beverages. Annals of Microbiology. 73: 37. [DOI: 10.1186/s13213-023-01740-9]
Arasu M.V., Jung M.-W., Ilavenil S., Jane M., Kim D.-H., Lee K.-D., Park H.-S., Hur T.-Y., Choi G.-J., Lim Y.-C., Al-Dhabi N.A., Choi K.-C. (2013). Isolation and characterization of antifungal compound from Lactobacillus plantarum KKC-10 from forage silage with potential beneficial properties. Journal of Applied Microbiology. 115: 1172-1185. [DOI: 10.1111/jam.12319]
Begley M., Hill C., Gahan C.G.M. (2006). Bile salt hydrolase activity in probiotics. Applied and Environmental Microbiology. 72: 1729-1738. [DOI: 10.1128/AEM.72.3.1729-1738.2006]
Beshiru A., Okareh O.T., Chigor V.N., Igbinosa E.O. (2018). Assessment of water quality of rivers that serves as water sources for drinking and domestic functions in rural and urban communities in Edo North, Nigeria. Environmental Monitoring and Assessment. 190:387. [DOI: 10.1007/s10661-018-6771-7]
Bin Masalam M.S., Bahieldin A., Alharbi M.G., Al-Masaudi S., Al-Jaouni S.K., Harakeh S.M., Al-Hindi R.R. (2018). Isolation, molecular characterization and probiotic potential of lactic acid bacteria in Saudi raw and fermented milk. Evidence-Based Complementary and Alternative Medicine. 2018: 7970463. [DOI: 10.1155/2018/7970463]
Caligiore-Gei P.F., Valdez J.G. (2015). Adjustment of a rapid method for the quantification of Fusarium spp. spore suspensions in plant pathology. Revista Argentina de Microbiología. 47: 152-154. [DOI: 10.1016/j.ram.2015.03.002]
Chavan P., Lata K., Kaur T., Rezek Jambrak A., Sharma S., Roy S., Sinhmar A., Thory R., Pal Singh G., Aayush K., Rout A. (2023). Recent advances in the preservation of postharvest fruits using edible films and coatings: a comprehensive review. Food Chemistry. 418: 135916. [DOI: 10.1016/j.foodchem. 2023.135916]
Cheesebrough M. (2006). District laboratory practice in tropical countries. 2nd edition. Cambridge University Press, Cambridge, UK. pp: 1-434. [DOI: 10.1017/CBO9780511543470]
Chen H., Ju H., Wang Y., Du G., Yan X., Cui Y., Yuan Y., Yue T. (2021). Antifungal activity and mode of action of lactic acid bacteria isolated from kefir against Penicillium expansum. Food Control. 130: 108274. [DOI: 10.1016/j.foodcont.2021.108274]
Chuku E.C. (2009). Fungi responsible for the spoilage of plantain (Musa paradisiaca) at different ripening stage. Acta Agronomica Nigeriana. 9: 35-40.
Crowley S., Mahony J., van Sinderen D. (2013). Current perspectives on antifungal lactic acid bacteria as natural bio-preservatives. Trends in Food Science and Technology. 33: 93-109. [DOI: 10.1016/j.tifs.2013.07.004]
Da Costa R.J., Voloski F.L.S., Mondadori R.G., Duval E.H., Fiorentini Â.M. (2019). Preservation of meat products with bacteriocins produced by lactic acid bacteria isolated from meat. Journal of Food Quality. 4726510. [DOI: 10.1155/2019/4726510]
Eni A.O., Oluwawemitan I.A., Solomon O.U. (2010). Microbial quality of fruits and vegetables sold in Sango Ota, Ogun State, Nigeria. African Journal of Food Science. 4: 291-296.
Erika M.T., Andrea K.B., Tamás F., Balázs V., Rita S., Károly M. (2013). Practical microbiology. Eötvös Loránd University, Budapest, Hungary. pp: 1-222. URL: https://ttk.elte.hu/dstore/document/893/book.pdf.
Hasegawa M., Kishino H., Yano T. (1985). Dating the human-ape split by a molecular clock of mitochondrial DNA. Journal of Molecular Evolution. 22: 160-174. [DOI: 10.1007/BF02101694]
Honfo F.G., Kayodé A.P.P., Coulibaly O., Tenkouano A. (2007). Relative contribution of banana and plantain products to the nutritional requirements for iron, zinc and vitamin A of infants and mothers in Cameroon. Fruits. 62: 267-277. [DOI: 10.1051/fruits:2007023]
Huan Y., Kong Q., Mou H., Yi H. (2020). Antimicrobial peptides: classification, design, application and research progress in multiple fields. Frontier in Microbiology. 11: 582779. [DOI: 10.3389/fmicb.2020.582779]
International Institute of Tropical Agriculture (IITA). (2000). Improving plantain and banana based. International Institute of Tropical Agriculture Annual Report. 13-14. URL: https:// www.iita.org/wp-content/uploads/2016/04/Annual-Report-2000-full-version.pdf.
Jeong S.-G., Kim H.M., Lee M., Yang J.E., Park H.W. (2023). Use of vegetable waste as a culture medium ingredient improves the antimicrobial and immunomodulatory activities of Lantiplantibacillus plantarum Wikim0125 isolated form kimchi. Journal of Microbiology and Biotechnology 33: 75-82. [DOI: 10.4014/jmb.2210.10049]
Kivanc M., Kivanc S.A., Pektas S. (2014). Screening of lactic acid bacteria for antifungal activity against fungi. Journal of Food Processing and Technology. 5: 1-4. [DOI: 10.4172/2157-7110.1000310]
Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution. 35: 1547-1549. [DOI: 10.1093/molbev/msy096]
Lahiri D., Nag M., Sarkar T., Ray R.R., Shariati M.A., Rebezov M., Bangar S.P., Lorenzo J.M., Domínguez R. (2021). Lactic Acid Bacteria (LAB): autochthonous and probiotic microbes for meat preservation and fortification. Foods. 11: 2792. [DOI: 10.3390/foods11182792]
Liu J., Huang R., Song Q., Xiong H., Ma J., Xia R., Qiao J. (2021). Combinational antibacterial activity of nisin and 3-phenyllactic acid and their co-production by engineered lactococcus lactis. Frontiers in Bioengineering and Biotechnology. 9: 612105. [DOI: 10.3389/fbioe.2021.612105]
Luz C., Saladino F., Lusiano F.B., Mañes J., Meca G. (2017). Invitro antifungal activity of bioactive peptides produced by Lactobacillus plantarum against Aspergillus parasiticus and Penicillium expansum. LWT - Food Science and Technology. 81: 128-135. [DOI: 10.1016/j.lwt.2017.03.053]
Ma H., Difazio S. (2008). An efficient method for purification of PCR products for sequencing. Biotechniques. 44: 921-923. [DOI: 10.2144/000112809]
Mafe A.N., Edo G.I., Makia R.S., Joshua O.A., Akpoghelie P.O., Gaaz T.S., Jikah A.N., Yousif E., Isoje E.F., Igbuku U.A., Ahmed D.S., Essaghah A.E.A., et al. (2024). A review on food spoilage mechanisms, food borne diseases and commercial aspects of food preservation and processing. Food Chemistry Advances. 5: 100852. [DOI: 10.1016/j.focha.2024.100852]
Mani-López E., Arrioja-Bretón D., López-Malo A. (2022). The impacts of antimicrobial and antifungal activity of cell-free supernatants from lactic acid bacteria in vitro and foods. Comprehensive Reviews in Food Science and Food Safety. 21: 604-641. [DOI: 10.1111/1541-4337.12872]
Miedes E., Lorences E.P. (2004). Apple (Malus domestica) and tomato (Lycopersicum esculentum) fruits cell wall hemicelluloses and xyloglucan degradation during Penicillium expansum infection. Journal of Agriculture and Food Chemistry. 52: 7957-7963. [DOI: 10.1021/jf048890f]
Mogaji K.J., Arotupin D.J., Mogaji O., Arogunjo A.O., Ajayi-Moses O.B., Akinwunmi I.M., Gabriel P.O., Orekoya E.S., Olajesu O.L., Adeleye H.J., Aribisala J.O. (2021). Inherent microorganisms affects the quality of a Nieria fermented beverage “Agadagidi” during production. Asian Food Science Journal. 20: 6-17. [DOI: 10.9734/afsj/2021/v20i730316]
Ofosu D.O., Before I., Martinson F., Frimpong G.K., Asare I.K., Darfour B. (2023). Pre-harvest and post-harvest practices along the plantain (Musa spp. AAB) fruit value chain in Ghana that predispose them to ripening. Ghana Journal of Agricultural Science. 58: 75–82.
Okafor S.E., Eni A.O. (2018). Microbial quality and the occurrence of aflatoxins in plantain/yam and wheat flours in Ado-Odo Ota. Earth and Environmental Science. 210: 1-22. [DOI: 10.1088/1755-1315/210/1/012017]
Olutiola P.O., Famurewa, O., Suntay, H.G. (1991). An introduction to general microbiology: a practical approach. Hygiene Institut der Universitat Heidelberg. Heidelberg. URL: https://www.scirp.org/reference/referencespapers?referenceid=561804.
Onipede G., Aremu B., Sanni A., Babalola O. (2020). Molecular study of the phytase gene in lactic acid bacteria isolated from Ogi and Kunun-zaki, African fermented cereal gruel and beverage. Applied Food Biotechnology. 7: 49-60. [DOI: 10.22037/afb.v7i1.25909]
Oriola O., Boboye B., Adetuyi O. (2017). Microorganisms associated with the production of a Nigerian fermented beverage, ‘Agadagidi’. Microbiology Research Journal International. 20: 1-9. [DOI: 10.9734/MRJI/2017/32654]
Oyewole O.F., Maria C.O., Tope P.S., Funmi O.O. (2018). In vitro study of potential probiotic lactic acid bacteria isolated from the gut of chickens in Abeokuta, Nigeria. Alexandria Journal of Veterinary Science. 58: 73-84. [DOI: 10.5455/ajvs.290499]
Petrikkou E., Rodrı́guez-Tudela J.L., Cuenca-Estrella M., Gomez A., Molleja A., Mellado E. (2001). Inoculum standardization for antifungal susceptibility testing of filamentous fungi pathogenic for humans. Journal of Clinical Microbiology. 39: 1345-1347. [DOI: 10.1128/JCM.39.4.1345-1347.2001]
Ramudingana P., Mamphogoro T.P., Kamutando C.N., Maboko M.M., Modika K.Y., Moloto K.W., Thantsha M.S. (2024). Antagonistic potential of endophytic fungal isolates of tomato (Solanum lycopersicum L.) fruits against post-harvest disease-causing pathogens of tomatoes: An in vitro investigation. Fungal Biology. 128: 1847-1858. [DOI: 10.1016/j.funbio.2024.05.006]
Rios Covian D., Ruas-Madiedo P., Margolles A., Gueimonde M., De Los Reyes- Gavilán C.G., Salazar N. (2016). Intestinal short chain fatty acids and their link with diet and human health. Frontiers in Microbiology. 7: 185. [DOI: 10.3389/fmicb. 2016.00185]
Robinson J.C., Anderson T., Eckstein K. (1992). The influence of functional leaf removal at flower emergence on components of yield and photosynthetic compensation in banana. Journal of Horticultural Science. 67: 403-410. [DOI: 10.1080/00221589. 1992.11516265]
Rose-Monde M., Séverin K.K.A., Lessoy Z., Edwige A., Koffi Y., Sébastien N.L. (2013). Microbiological safety and quality assessment of a very appreciate traditional ready to eat plantain food, sold in retails markets. African Journal of Microbiology Research. 7: 4123-4129.
Rouse S., Harnett D., Vaughan A., Van Sinderen D. (2008). Lactic acid bacteria with potential to eliminate fungal spoilage in foods. Journal of Applied Microbiology. 104: 915-923. [DOI: 10.1111/j.1365-2672.2007.03619.x]
Salas M.L., Mounier J., Valence F., Coton M., Thierry A., Coton E. (2017). Antifungal microbial agents for food biopreservation—a review. Microorganisms. 5: 37. [DOI: 10.3390/ microorganisms5030037]
Shehata M.G., Badr A.N., El Sohaimy S.A., Asker D., Awad T.S. (2019). Characterization of antifungal metabolites produced by novel lactic acid bacterium and their potential application as food biopreservatives. Annals of Agricultural Sciences. 64: 71-78. [DOI: 10.1016/j.aoas.2019.05.002]
Silva C.C.G., Silva S.P.M., Ribeiro S.C. (2018). Application of bacteriocins and protective cultures in dairy food preservation. Frontier in Microbiology. 9: 594. [DOI: 10.3389/fmicb.2018.00594]
Sofo A., Elshafie H.S., Camele I. (2020). Structural and functional organization of the root system: a comparative study on five plant species. Plants. 9: 1338. [DOI: 10.3390/plants9101338]
Strano M.C., Altieri G., Allergra M., Di Renzo G.C., Paterna G., Matera A., Genovese F. (2022). Postharvest technologies of fresh citrus fruit: advances and recent developments for loss reduction during handling and storage. Horticulturae. 8: 612. [DOI: 10.3390/horticulturae8070612]
Temilade O.P., Olubukola O.J., Akinkunmi W.A. Rukayat H.O. (2020). Antibacterial sensitivity profile of Lactobacillus sp and characterization of aflatoxin-producing fungi isolated from yam and plantain flour. The International Journal of Biotechnology. 9: 14-23. [DOI: 10.18488/journal.57.2020.91.14.23]
Udoh I.P., Eleazar C.I., Ogeneh B.O., Ohanu M.E. (2015). Studies on fungi responsible for the spoilage/deterioration of some edible fruits and vegetables. Advances in Microbiology. 5: 285-290. [DOI: 10.4236/aim.2015.54027]
Vida C., Evelyn C.I., Akrofi-Ansah H., Edwina A., Beatrice Q. (2024). Food processing: reducing postharvest losses of plantain through alternative recipe development. Acta Scientific Nutritional Health. 8: 60-66.
Wang H., Yan Y., Wang J., Zhang H., Qi W. (2012). Production and characterization of antifungal compounds produced by Lactobacillus plantarum IMAU10014. PLOS ONE. 7: e29452. [DOI: 10.1371/journal.pone.0029452]
Zanotti I., Turroni F., Piemontese A., Mancabelli L., Milani C., Viappiani A., Prevedini G., Sanchez B., Margolles A., Elviri L., Franco B., Van Sinderen D., et al. (2015). Evidence for cholesterol lowering activity by Bifidobacterium bifidum PRL2010 through gut microbiota modulation. Applied Microbiology and Biotechnology. 99: 6813-6829. [DOI: 10.1007/s00253-015-6564-7]
Zapaśnik A., Sokołowska B., Bryła M. (2022). Role of lactic acid bacteria in food preservation and safety. Foods. 11: 1283. [DOI: 10.3390/foods11091283]
Zebboudj N., Yezli W., Hamini-Kadar N., Kihal M., Henni J.E. (2014). Antifungal activity of lactic acid bacteria against Fusarium oxysporum f. sp. albedinis isolated from diseased date palm in South Algeria. International Journal of Biosciences. 5: 99-106. [DOI: 10.22364/eeb.18.02]
*Corresponding author (G.O. Onipede)
* E-mail: gbemisola.onipede@fuhsi.edu.ng
ORCID ID: https://orcid.org/0000-0003-3953-6104
* E-mail: gbemisola.onipede@fuhsi.edu.ng
ORCID ID: https://orcid.org/0000-0003-3953-6104
Type of Study: Original article |
Subject:
Special
Received: 24/08/11 | Accepted: 25/08/25 | Published: 25/09/30
Received: 24/08/11 | Accepted: 25/08/25 | Published: 25/09/30
References
1. Abdelkader M., Chihib N.E., Abdelkader D.B., Naima N.A., Jean P.H. (2009). Antibacterial activity of some lactic acid bacteria isolated from an Algerian dairy product. Journal of Environmental and Public Health. 1-6. [DOI: 10.1155/2009/678495] [DOI:10.1155/2009/678495] [PMID] [PMCID]
2. Abdu M.M., Rashid S., Beatrice K. (2024). Determination of aflatoxin level in stored maize flour and risk of exposure from consumption of aflatoxin contaminated meal in boarding school. International Journal of Microbiology and Biotechnology. 9: 7-14. [DOI: 10.11648/ijmb.20240901.12] [DOI:10.11648/ijmb.20240901.12]
3. Abiodun-Solanke A.O., Falade K.O. (2010). A review of the uses and methods of processing banana and plantain (Musa spp.) into storable food products. Journal of Agricultural Research and Development. 9. [DOI: 10.4314/jard.v9i2.66815] [DOI:10.4314/jard.v9i2.66815]
4. Ajayi A.O. (2016). Microbiological quality of plantain (musa paradisiacal). Nigerian Journal of Microbiology. 30: 3611-3618.
5. Al-Hindi R.R., Al-Najada A.R., Mohamed S.A. (2011). Isolation and identification of some fruit spoilage fungi: screening of plant cell wall degrading enzymes. African Journal of Microbiology Research. 5: 443-448. [DOI: 10.5897/AJMR10.896]
6. Amenu D., Bacha K. (2023). Probiotic potential and safety analysis of lactic acid bacteria isolated from Ethiopian traditional fermented foods and beverages. Annals of Microbiology. 73: 37. [DOI: 10.1186/s13213-023-01740-9] [DOI:10.1186/s13213-023-01740-9]
7. Arasu M.V., Jung M.-W., Ilavenil S., Jane M., Kim D.-H., Lee K.-D., Park H.-S., Hur T.-Y., Choi G.-J., Lim Y.-C., Al-Dhabi N.A., Choi K.-C. (2013). Isolation and characterization of antifungal compound from Lactobacillus plantarum KKC-10 from forage silage with potential beneficial properties. Journal of Applied Microbiology. 115: 1172-1185. [DOI: 10.1111/jam.12319] [DOI:10.1111/jam.12319] [PMID]
8. Begley M., Hill C., Gahan C.G.M. (2006). Bile salt hydrolase activity in probiotics. Applied and Environmental Microbiology. 72: 1729-1738. [DOI: 10.1128/AEM.72.3.1729-1738.2006] [DOI:10.1128/AEM.72.3.1729-1738.2006] [PMID] [PMCID]
9. Beshiru A., Okareh O.T., Chigor V.N., Igbinosa E.O. (2018). Assessment of water quality of rivers that serves as water sources for drinking and domestic functions in rural and urban communities in Edo North, Nigeria. Environmental Monitoring and Assessment. 190:387. [DOI: 10.1007/s10661-018-6771-7] [DOI:10.1007/s10661-018-6771-7] [PMID]
10. Bin Masalam M.S., Bahieldin A., Alharbi M.G., Al-Masaudi S., Al-Jaouni S.K., Harakeh S.M., Al-Hindi R.R. (2018). Isolation, molecular characterization and probiotic potential of lactic acid bacteria in Saudi raw and fermented milk. Evidence-Based Complementary and Alternative Medicine. 2018: 7970463. [DOI: 10.1155/2018/7970463] [DOI:10.1155/2018/7970463] [PMID] [PMCID]
11. Caligiore-Gei P.F., Valdez J.G. (2015). Adjustment of a rapid method for the quantification of Fusarium spp. spore suspensions in plant pathology. Revista Argentina de Microbiología. 47: 152-154. [DOI: 10.1016/j.ram.2015.03.002] [DOI:10.1016/j.ram.2015.03.002] [PMID]
12. Chavan P., Lata K., Kaur T., Rezek Jambrak A., Sharma S., Roy S., Sinhmar A., Thory R., Pal Singh G., Aayush K., Rout A. (2023). Recent advances in the preservation of postharvest fruits using edible films and coatings: a comprehensive review. Food Chemistry. 418: 135916. [DOI: 10.1016/j.foodchem. 2023.135916] [DOI:10.1016/j.foodchem.2023.135916] [PMID]
13. Cheesebrough M. (2006). District laboratory practice in tropical countries. 2nd edition. Cambridge University Press, Cambridge, UK. pp: 1-434. [DOI: 10.1017/CBO9780511543470] [DOI:10.1017/CBO9780511543470] [PMCID]
14. Chen H., Ju H., Wang Y., Du G., Yan X., Cui Y., Yuan Y., Yue T. (2021). Antifungal activity and mode of action of lactic acid bacteria isolated from kefir against Penicillium expansum. Food Control. 130: 108274. [DOI: 10.1016/j.foodcont.2021.108274] [DOI:10.1016/j.foodcont.2021.108274]
15. Chuku E.C. (2009). Fungi responsible for the spoilage of plantain (Musa paradisiaca) at different ripening stage. Acta Agronomica Nigeriana. 9: 35-40.
16. Crowley S., Mahony J., van Sinderen D. (2013). Current perspectives on antifungal lactic acid bacteria as natural bio-preservatives. Trends in Food Science and Technology. 33: 93-109. [DOI: 10.1016/j.tifs.2013.07.004] [DOI:10.1016/j.tifs.2013.07.004]
17. Da Costa R.J., Voloski F.L.S., Mondadori R.G., Duval E.H., Fiorentini Â.M. (2019). Preservation of meat products with bacteriocins produced by lactic acid bacteria isolated from meat. Journal of Food Quality. 4726510. [DOI: 10.1155/2019/4726510] [DOI:10.1155/2019/4726510]
18. Eni A.O., Oluwawemitan I.A., Solomon O.U. (2010). Microbial quality of fruits and vegetables sold in Sango Ota, Ogun State, Nigeria. African Journal of Food Science. 4: 291-296.
19. Erika M.T., Andrea K.B., Tamás F., Balázs V., Rita S., Károly M. (2013). Practical microbiology. Eötvös Loránd University, Budapest, Hungary. pp: 1-222. URL: https://ttk.elte.hu/dstore/document/893/book.pdf.
20. Hasegawa M., Kishino H., Yano T. (1985). Dating the human-ape split by a molecular clock of mitochondrial DNA. Journal of Molecular Evolution. 22: 160-174. [DOI: 10.1007/BF02101694] [DOI:10.1007/BF02101694] [PMID]
21. Honfo F.G., Kayodé A.P.P., Coulibaly O., Tenkouano A. (2007). Relative contribution of banana and plantain products to the nutritional requirements for iron, zinc and vitamin A of infants and mothers in Cameroon. Fruits. 62: 267-277. [DOI:10.1051/fruits:2007023] [DOI:10.1051/fruits:2007023]
22. Huan Y., Kong Q., Mou H., Yi H. (2020). Antimicrobial peptides: classification, design, application and research progress in multiple fields. Frontier in Microbiology. 11: 582779. [DOI: 10.3389/fmicb.2020.582779] [DOI:10.3389/fmicb.2020.582779] [PMID] [PMCID]
23. International Institute of Tropical Agriculture (IITA). (2000). Improving plantain and banana based. International Institute of Tropical Agriculture Annual Report. 13-14. URL: https:// www.iita.org/wp-content/uploads/2016/04/Annual-Report-2000-full-version.pdf.
24. Jeong S.-G., Kim H.M., Lee M., Yang J.E., Park H.W. (2023). Use of vegetable waste as a culture medium ingredient improves the antimicrobial and immunomodulatory activities of Lantiplantibacillus plantarum Wikim0125 isolated form kimchi. Journal of Microbiology and Biotechnology 33: 75-82. [DOI: 10.4014/jmb.2210.10049] [DOI:10.4014/jmb.2210.10049] [PMID] [PMCID]
25. Kivanc M., Kivanc S.A., Pektas S. (2014). Screening of lactic acid bacteria for antifungal activity against fungi. Journal of Food Processing and Technology. 5: 1-4. [DOI: 10.4172/2157-7110.1000310] [DOI:10.4172/2157-7110.1000310]
26. Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution. 35: 1547-1549. [DOI: 10.1093/molbev/msy096] [DOI:10.1093/molbev/msy096] [PMID] [PMCID]
27. Lahiri D., Nag M., Sarkar T., Ray R.R., Shariati M.A., Rebezov M., Bangar S.P., Lorenzo J.M., Domínguez R. (2021). Lactic Acid Bacteria (LAB): autochthonous and probiotic microbes for meat preservation and fortification. Foods. 11: 2792. [DOI: 10.3390/foods11182792] [DOI:10.3390/foods11182792] [PMID] [PMCID]
28. Liu J., Huang R., Song Q., Xiong H., Ma J., Xia R., Qiao J. (2021). Combinational antibacterial activity of nisin and 3-phenyllactic acid and their co-production by engineered lactococcus lactis. Frontiers in Bioengineering and Biotechnology. 9: 612105. [DOI: 10.3389/fbioe.2021.612105] [DOI:10.3389/fbioe.2021.612105] [PMID] [PMCID]
29. Luz C., Saladino F., Lusiano F.B., Mañes J., Meca G. (2017). Invitro antifungal activity of bioactive peptides produced by Lactobacillus plantarum against Aspergillus parasiticus and Penicillium expansum. LWT - Food Science and Technology. 81: 128-135. [DOI: 10.1016/j.lwt.2017.03.053] [DOI:10.1016/j.lwt.2017.03.053]
30. Ma H., Difazio S. (2008). An efficient method for purification of PCR products for sequencing. Biotechniques. 44: 921-923. [DOI: 10.2144/000112809] [DOI:10.2144/000112809] [PMID]
31. Mafe A.N., Edo G.I., Makia R.S., Joshua O.A., Akpoghelie P.O., Gaaz T.S., Jikah A.N., Yousif E., Isoje E.F., Igbuku U.A., Ahmed D.S., Essaghah A.E.A., et al. (2024). A review on food spoilage mechanisms, food borne diseases and commercial aspects of food preservation and processing. Food Chemistry Advances. 5: 100852. [DOI: 10.1016/j.focha.2024.100852] [DOI:10.1016/j.focha.2024.100852]
32. Mani-López E., Arrioja-Bretón D., López-Malo A. (2022). The impacts of antimicrobial and antifungal activity of cell-free supernatants from lactic acid bacteria in vitro and foods. Comprehensive Reviews in Food Science and Food Safety. 21: 604-641. [DOI: 10.1111/1541-4337.12872] [DOI:10.1111/1541-4337.12872] [PMID]
33. Miedes E., Lorences E.P. (2004). Apple (Malus domestica) and tomato (Lycopersicum esculentum) fruits cell wall hemicelluloses and xyloglucan degradation during Penicillium expansum infection. Journal of Agriculture and Food Chemistry. 52: 7957-7963. [DOI: 10.1021/jf048890f] [DOI:10.1021/jf048890f] [PMID]
34. Mogaji K.J., Arotupin D.J., Mogaji O., Arogunjo A.O., Ajayi-Moses O.B., Akinwunmi I.M., Gabriel P.O., Orekoya E.S., Olajesu O.L., Adeleye H.J., Aribisala J.O. (2021). Inherent microorganisms affects the quality of a Nieria fermented beverage "Agadagidi" during production. Asian Food Science Journal. 20: 6-17. [DOI: 10.9734/afsj/2021/v20i730316] [DOI:10.9734/afsj/2021/v20i730316]
35. Ofosu D.O., Before I., Martinson F., Frimpong G.K., Asare I.K., Darfour B. (2023). Pre-harvest and post-harvest practices along the plantain (Musa spp. AAB) fruit value chain in Ghana that predispose them to ripening. Ghana Journal of Agricultural Science. 58: 75-82. [DOI:]
36. Okafor S.E., Eni A.O. (2018). Microbial quality and the occurrence of aflatoxins in plantain/yam and wheat flours in Ado-Odo Ota. Earth and Environmental Science. 210: 1-22. [DOI: 10.1088/1755-1315/210/1/012017] [DOI:10.1088/1755-1315/210/1/012017]
37. Olutiola P.O., Famurewa, O., Suntay, H.G. (1991). An introduction to general microbiology: a practical approach. Hygiene Institut der Universitat Heidelberg. Heidelberg. URL: https://www.scirp.org/reference/referencespapers?referenceid=561804.
38. Onipede G., Aremu B., Sanni A., Babalola O. (2020). Molecular study of the phytase gene in lactic acid bacteria isolated from Ogi and Kunun-zaki, African fermented cereal gruel and beverage. Applied Food Biotechnology. 7: 49-60. [DOI: 10.22037/afb.v7i1.25909]
39. Oriola O., Boboye B., Adetuyi O. (2017). Microorganisms associated with the production of a Nigerian fermented beverage, 'Agadagidi'. Microbiology Research Journal International. 20: 1-9. [DOI: 10.9734/MRJI/2017/32654] [DOI:10.9734/MRJI/2017/32654]
40. Oyewole O.F., Maria C.O., Tope P.S., Funmi O.O. (2018). In vitro study of potential probiotic lactic acid bacteria isolated from the gut of chickens in Abeokuta, Nigeria. Alexandria Journal of Veterinary Science. 58: 73-84. [DOI: 10.5455/ajvs.290499] [DOI:10.5455/ajvs.290499]
41. Petrikkou E., Rodrı́guez-Tudela J.L., Cuenca-Estrella M., Gomez A., Molleja A., Mellado E. (2001). Inoculum standardization for antifungal susceptibility testing of filamentous fungi pathogenic for humans. Journal of Clinical Microbiology. 39: 1345-1347. [DOI: 10.1128/JCM.39.4.1345-1347.2001] [DOI:10.1128/JCM.39.4.1345-1347.2001] [PMID] [PMCID]
42. Ramudingana P., Mamphogoro T.P., Kamutando C.N., Maboko M.M., Modika K.Y., Moloto K.W., Thantsha M.S. (2024). Antagonistic potential of endophytic fungal isolates of tomato (Solanum lycopersicum L.) fruits against post-harvest disease-causing pathogens of tomatoes: An in vitro investigation. Fungal Biology. 128: 1847-1858. [DOI: 10.1016/j.funbio.2024.05.006] [DOI:10.1016/j.funbio.2024.05.006] [PMID]
43. Rios Covian D., Ruas-Madiedo P., Margolles A., Gueimonde M., De Los Reyes- Gavilán C.G., Salazar N. (2016). Intestinal short chain fatty acids and their link with diet and human health. Frontiers in Microbiology. 7: 185. [DOI: 10.3389/fmicb. 2016.00185] [DOI:10.3389/fmicb.2016.00185] [PMID] [PMCID]
44. Robinson J.C., Anderson T., Eckstein K. (1992). The influence of functional leaf removal at flower emergence on components of yield and photosynthetic compensation in banana. Journal of Horticultural Science. 67: 403-410. [DOI: 10.1080/00221589. 1992.11516265] [DOI:10.1080/00221589.1992.11516265]
45. Rose-Monde M., Séverin K.K.A., Lessoy Z., Edwige A., Koffi Y., Sébastien N.L. (2013). Microbiological safety and quality assessment of a very appreciate traditional ready to eat plantain food, sold in retails markets. African Journal of Microbiology Research. 7: 4123-4129.
46. Rouse S., Harnett D., Vaughan A., Van Sinderen D. (2008). Lactic acid bacteria with potential to eliminate fungal spoilage in foods. Journal of Applied Microbiology. 104: 915-923. [DOI: 10.1111/j.1365-2672.2007.03619.x] [DOI:10.1111/j.1365-2672.2007.03619.x] [PMID]
47. Salas M.L., Mounier J., Valence F., Coton M., Thierry A., Coton E. (2017). Antifungal microbial agents for food biopreservation-a review. Microorganisms. 5: 37. [DOI: 10.3390/ microorganisms5030037] [DOI:10.3390/microorganisms5030037] [PMID] [PMCID]
48. Shehata M.G., Badr A.N., El Sohaimy S.A., Asker D., Awad T.S. (2019). Characterization of antifungal metabolites produced by novel lactic acid bacterium and their potential application as food biopreservatives. Annals of Agricultural Sciences. 64: 71-78. [DOI: 10.1016/j.aoas.2019.05.002] [DOI:10.1016/j.aoas.2019.05.002]
49. Silva C.C.G., Silva S.P.M., Ribeiro S.C. (2018). Application of bacteriocins and protective cultures in dairy food preservation. Frontier in Microbiology. 9: 594. [DOI: 10.3389/fmicb.2018.00594] [DOI:10.3389/fmicb.2018.00594] [PMID] [PMCID]
50. Sofo A., Elshafie H.S., Camele I. (2020). Structural and functional organization of the root system: a comparative study on five plant species. Plants. 9: 1338. [DOI: 10.3390/plants9101338] [DOI:10.3390/plants9101338] [PMID] [PMCID]
51. Strano M.C., Altieri G., Allergra M., Di Renzo G.C., Paterna G., Matera A., Genovese F. (2022). Postharvest technologies of fresh citrus fruit: advances and recent developments for loss reduction during handling and storage. Horticulturae. 8: 612. [DOI: 10.3390/horticulturae8070612] [DOI:10.3390/horticulturae8070612]
52. Temilade O.P., Olubukola O.J., Akinkunmi W.A. Rukayat H.O. (2020). Antibacterial sensitivity profile of Lactobacillus sp and characterization of aflatoxin-producing fungi isolated from yam and plantain flour. The International Journal of Biotechnology. 9: 14-23. [DOI: 10.18488/journal.57.2020.91.14.23] [DOI:10.18488/journal.57.2020.91.14.23]
53. Udoh I.P., Eleazar C.I., Ogeneh B.O., Ohanu M.E. (2015). Studies on fungi responsible for the spoilage/deterioration of some edible fruits and vegetables. Advances in Microbiology. 5: 285-290. [DOI: 10.4236/aim.2015.54027] [DOI:10.4236/aim.2015.54027]
54. Vida C., Evelyn C.I., Akrofi-Ansah H., Edwina A., Beatrice Q. (2024). Food processing: reducing postharvest losses of plantain through alternative recipe development. Acta Scientific Nutritional Health. 8: 60-66.
55. Wang H., Yan Y., Wang J., Zhang H., Qi W. (2012). Production and characterization of antifungal compounds produced by Lactobacillus plantarum IMAU10014. PLOS ONE. 7: e29452. [DOI: 10.1371/journal.pone.0029452] [DOI:10.1371/journal.pone.0029452] [PMID] [PMCID]
56. Zanotti I., Turroni F., Piemontese A., Mancabelli L., Milani C., Viappiani A., Prevedini G., Sanchez B., Margolles A., Elviri L., Franco B., Van Sinderen D., et al. (2015). Evidence for cholesterol lowering activity by Bifidobacterium bifidum PRL2010 through gut microbiota modulation. Applied Microbiology and Biotechnology. 99: 6813-6829. [DOI: 10.1007/s00253-015-6564-7] [DOI:10.1007/s00253-015-6564-7] [PMID]
57. Zapaśnik A., Sokołowska B., Bryła M. (2022). Role of lactic acid bacteria in food preservation and safety. Foods. 11: 1283. [DOI: 10.3390/foods11091283] [DOI:10.3390/foods11091283] [PMID] [PMCID]
58. Zebboudj N., Yezli W., Hamini-Kadar N., Kihal M., Henni J.E. (2014). Antifungal activity of lactic acid bacteria against Fusarium oxysporum f. sp. albedinis isolated from diseased date palm in South Algeria. International Journal of Biosciences. 5: 99-106. [DOI: 10.22364/eeb.18.02] [DOI:10.22364/eeb.18.02]
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |