Volume 12, Issue 3 (September 2025)
J. Food Qual. Hazards Control 2025, 12(3): 192-200 |
Back to browse issues page
Ethics code: Not applicable.
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Balla B, Amoikon T, Aka-Gbezo S, N’Sa K, Kamagate M, Tié L et al . Molecular Characterization of Tamarindus indica Associated with Fungal Diversity and Detection of Aflatoxin Biosynthesiser Genes. J. Food Qual. Hazards Control 2025; 12 (3) :192-200
URL: http://jfqhc.ssu.ac.ir/article-1-1319-en.html
URL: http://jfqhc.ssu.ac.ir/article-1-1319-en.html
Unité de Formation et de Recherche en Sciences et Technologie des Aliments (UFR-STA), Université Nangui Abrogoua , Abidjan, Côte d’Ivoire, Centre Suisse de Recherches Scientifiques en Côte d’Ivoire, Abidjan, Côte d’Ivoire , solange.aka@csrs.ci
Keywords: Tamarindus, Polymorphism, Genetic, Restriction Fragment Length, Aspergillus, Aflatoxins, Food Contamination
Full-Text [PDF 506 kb]
(208 Downloads)
| Abstract (HTML) (438 Views)
Full-Text: (70 Views)
Molecular Characterization of Tamarindus indica Associated with Fungal Diversity and Detection of Aflatoxin Biosynthesiser Genes
B. Balla [1], T.L.S. Amoikon 2,3, S. Aka-Gbezo 1,3** , K.M.C N’Sa 1, M. Kamagate 1,
L.I.A. Tié 1, M.K. Djé 1
1. Unité de Formation et de Recherche en Sciences et Technologie des Aliments (UFR-STA), Université Nangui Abrogoua , Abidjan, Côte d’Ivoire
2. Institut Pierre Richet, Institut National de Santé Publique, Bouaké, Côte d’Ivoire
3. Centre Suisse de Recherches Scientifiques en Côte d’Ivoire, Abidjan, Côte d’Ivoire
HIGHLIGHTS
Table 1: Physicochemical properties of tamarind pulps
Table 2: Macroscopic and microscopic appearances of tamarind fungal isolates
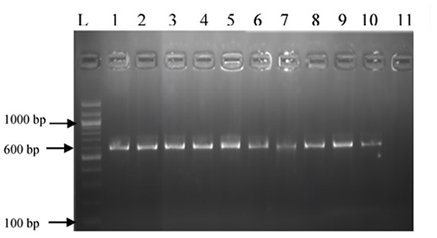
Figure 1: Polymerase Chain Reaction (PCR) product of the Internal Transcribed Spacer (ITS-5.8S) rDNA region. L: 100 bp DNA ladder; lanes 1-10: PCR products from 10 out of 470 fungal isolates; lane 11: water control. The gel shows a 600 bp PCR product corresponding to the ITS-5.8S rDNA region of the fungal isolates
Enzymatic digestion using HaeIII, EcoRI, Sau3AI, and BslI produced fragments of 350 and 80 bp, 310 and 280 bp, 320 and 250 bp, and 435 and 90 bp, respectively (Table 3). BLAST analysis of the 5.8S-ITS sequence identified the species as Aspergillus melleus (99.12-100% identity) from the subgenus Circumdati and section Circumdati, with a prevalence of 20%. The identification of A. melleus highlights its potential role as a tamarind contaminant. While no pathogenic potential is commonly attributed to this species, its adaptation to low-moisture environments may explain its presence in tamarind samples. Group II exhibited powdery green colonies with a white margin and pale pink reverse. Microscopically, it displayed colorless, septate, branched mycelium, and biseriate conidial heads with spherical vesicles. Enzymatic digestion revealed fragments of 260, 110, and 70 bp for HaeIII; 295 and 280 bp for EcoRI; 330 and 180 bp for Sau3AI; and 350 and 160 bp for BslI. This group was identified as Monascus pilosus (99.65-100% identity), a species from the genus Monascus within the family Trichocomaceae, with a prevalence of 15%. Although M. pilosus is not commonly associated with mycotoxin production, its presence highlights the diversity of fungal species capable of contaminating tamarind. Group III showed flat gray-yellow colonies with colorless reverse and small forms. Microscopically, it exhibited colorless, septate mycelium, and brush-like ramified conidial heads. Fragments of 290, 100, and 70 bp for HaeIII; 300 and 290 bp for EcoRI; 230, 150, and 90 bp for Sau3AI; and 380 and 180 bp for BslI were observed. BLAST analysis confirmed the species as Aspergillus aflatoxiformans (100% identity) (Table 3) from the section Flavi, with a prevalence of 5%. A. aflatoxiformans has been identified as a producer of aflatoxins B1, B2, G1, and G2 and is commonly detected in foods across Africa (Frisvad et al., 2019). Among the major mycotoxins, aflatoxins, classified as furanocoumarin-derived polyketides, are considered to be the most toxic and carcinogenic. Aflatoxins are commonly found contaminating various agricultural products and present significant health risks to both humans and livestock (Navale et al., 2021). Group IV showed granular black colonies with a white margin and beige reverse. It featured septate, branched mycelium, a fast-growing thallus, globose brown conidia, and slightly flattened vesicles. The restriction fragments obtained were 290, 90 and 70 bp for HaeIII; 300 and 290 bp for EcoRI; 230, 210 and 90 bp for Sau3AI; and 300, 180, and 70 bp for BslI. The species was identified as Aspergillus niger (99.62-100% identity) from the section Nigri, with a prevalence of 20%. A. niger is known for its ability to produce ochratoxin A (OTA), on natural substrates, although species within the Nigri section generally produce lower levels of this toxin. OTA is commonly formed during storage of crops and has been identified as neurotoxic, hepatotoxic, immunotoxic, genotoxic, embryotoxic, teratogenic, and carcinogenic in both humans and animals (Navale et al., 2021). Group V displayed powdery brown colonies spreading on the medium with a beige reverse. Microscopically, it featured septate, branched mycelium, and globose conidiophores. This group exhibited restriction patterns identical to Group IV. The species was identified as Aspergillus costaricensis (99.67–100% identity) from the section Nigri, with a prevalence of 40%.
A recent study by Bian et al. (2022) reclassified 14 Aspergillus species within the Nigri section into six species, with A. costaricensis being reclassified as Aspergillus tubingensis. The work of Kizis et al. (2014) showed that the polymorphism between A. niger and A. tubingensis could be distinguished using the enzyme RsaI. Therefore, A. niger and A. costaricensis could potentially be distinguished in our study using the RsaI enzyme. However, it should be noted that no previous study has documented the RFLP profiles of the fungi analyzed here. Therefore, this study contributes to the enrichment of existing RFLP profile databases for fungi and underscores the utility of combining morphological identification, RFLP profiling, and sequencing for fungal species characterization. While RFLP is cost-effective for reducing sequencing needs, its limitations in distinguishing closely related species, such as A. niger and A. costaricensis, were evident. The prominence of Aspergillus species across tamarind samples can be attributed to their ability to thrive in low-moisture environments, as observed in grains where moisture levels below 13% did not significantly inhibit fungal growth (Nji et al., 2022). The detection of mycotoxin-producing species, such as A. niger and A. aflatoxiformans, highlights potential health risks and underscores the importance of storage and quality control measures for tamarind products.
Table 3: Fungal identities and restriction profiles
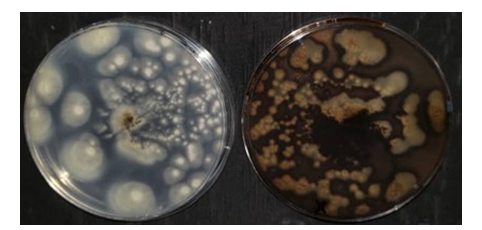
Figure 2: Qualitative screening of aflatoxin production in Aspergillus aflatoxiformans plates exposed to ammonia vapor (Left: beige control, Right: color change to plum red in isolate)
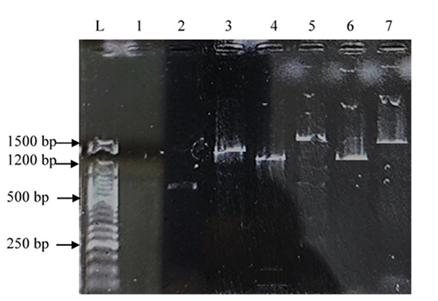
Figure 3: Polymerase Chain Reaction (PCR) product targeting the aflR gene in Aspergillus aflatoxiformans. L: 50 bp DNA Ladder; Lane 1: aflD gene; Lane 2: aflQ gene; Lane 3: aflM gene; Lane 4: aflP gene; Lane 5: aflR gene; Lane 6: aflO gene
B. Balla [1], T.L.S. Amoikon 2,3, S. Aka-Gbezo 1,3**
L.I.A. Tié 1, M.K. Djé 1
1. Unité de Formation et de Recherche en Sciences et Technologie des Aliments (UFR-STA), Université Nangui Abrogoua , Abidjan, Côte d’Ivoire
2. Institut Pierre Richet, Institut National de Santé Publique, Bouaké, Côte d’Ivoire
3. Centre Suisse de Recherches Scientifiques en Côte d’Ivoire, Abidjan, Côte d’Ivoire
- Tamarind's acidity and low moisture enhanced its shelf stability.
- Aspergillus aflatoxiformans carried genes for aflatoxin biosynthesis.
- RFLP grouped the fungi but lacked species-level resolution.
| Article type Original article |
ABSTRACT Background: The tamarind tree (Tamarindus indica), valued for its nutritional and economic importance, faces significant food safety challenges due to fungal contamination and mycotoxin production. This study aims to characterize the toxigenic fungal flora of tamarind pulp to provide crucial information for developing improved management practices to ensure food safety and protect public health in the region. Methods: Tamarind samples (n=180) collected from different locations were analyzed for pH, Titratable Acidity, and Moisture Content. Fungal diversity was assessed with emphasis on the molecular identification of Aspergillus species. The ammonia vapor test was used to detect aflatoxigenic isolates, while the presence of key aflatoxin biosynthesis genes (aflD, aflM, aflO, aflP, aflQ, and aflR) was investigated by Polymerase Chain Reaction amplification. Restriction Fragment Length Polymorphism analysis was applied to group isolates. The data were analyzed using the Kruskal-Wallis test in R software (version 4.4.3; R Foundation for Statistical Computing) to assess differences between groups. Statistical significance was determined at p<0.05. Results: Tamarind was highly acidic (pH<3) with low Titratable Acidity (4-5%) and low water content (14-20%), which improves storage stability. Fungal analysis identified five species, namely Monascus pilosus, Aspergillus melleus, Aspergillus aflatoxiformans, Aspergillus niger, and Aspergillus costaricensis (the most frequently isolated species). Molecular analysis revealed amplification of four distinct fungal profiles, highlighting the usefulness of Restriction Fragment Length Polymorphisms for grouping isolates. However, closely related species such as A. niger and A. costaricensis were difficult to distinguish. Amplification of the aflD, aflM, aflO, aflP, aflQ, and aflR genes in A. aflatoxiformans isolates confirmed their ability to synthesise aflatoxins. Conclusion: This study highlighted the vulnerability of tamarind to contamination by aflatoxin-producing fungi. Monitoring mycotoxin levels and improving storage practices are essential to ensure tamarind safety and protect consumer health. © 2025, Shahid Sadoughi University of Medical Sciences. This is an open access article under the Creative Commons Attribution 4.0 International License. |
|
| Keywords Tamarindus Polymorphism Genetic Restriction Fragment Length Aspergillus Aflatoxins Food Contamination |
||
| Article history Received: 7 Jan 2025 Revised: 26 Apr 2025 Accept: 22 Jul 2025 |
||
| Abbreviations BLAST=Basic Local Alignment Search Tool DRBC=Dichloran Rose Bengale Chloramphénicol ITS=Internal Transcribed Spacer MC=Moisture Content PCR=Polymerase Chain Reaction RFLP=Restriction Fragment Length Polymorphism TA=Titratable Acidity |
To cite: Balla B., Amoikon T.L.S., Aka-Gbezo S., N’Sa K.M.C., Kamagate M., Tié L.I.A., Djé M.K. (2025). Molecular characterization of Tamarindus indica associated with fungal diversity and detection of aflatoxin biosynthesiser genes. Journal of Food Quality and Hazards Control. 12: 192-200.
Introduction
The tamarind tree Tamarindus indica, a monotypic species in the Fabaceae family, is native to tropical Africa. Its widespread cultivation in tropical regions (Diallo et al., 2010) highlights its importance across various sectors, including nutrition, traditional medicine, and cultural practices. In West Africa, tamarind plays a crucial role not only in traditional diets and therapeutic but also in the economic sustainability of local communities (Hamacek et al., 2013; Samarou et al., 2022a,b; Stege et al., 2011). Tamarind pulp is rich in macronutrients and micronutrients, particularly essential amino acids, and represents an excellent source of vitamin C. It is also rich in pectin and organic acids, particularly tartaric acid, and is extensively used in various food preparations such as sauces, jams, and fruit pastes (Bakayoko et al., 2024b; Devi and Boruah, 2020; Grollier et al., 1998; Hamacek et al., 2013; Samarou et al., 2022b). These culinary applications underscore not only the nutritional richness of tamarind pulp but also its versatility in local food traditions.
Despite its benefits, tamarind fruit faces significant challenges primarily due to its susceptibility to microbial contamination. Mold, prevalent in tropical environments, poses a substantial threat during the harvesting, storage, and processing stages of tamarind. This contamination not only affects the quality of the fruit but also poses serious food safety risks due to the potential production of mycotoxins. These secondary toxic metabolites produced by certain molds can lead to severe health issues including immune disorders, liver damage, and cancer (Kabak, 2009; Kouadio et al., 2014; Mafe and Büsselberg, 2024). Therefore, understanding the microbial flora associated with tamarind fruit, especially molds capable of producing mycotoxins, is crucial for ensuring food safety.
In Côte d'Ivoire, tamarind pulp, often more or less dried, is used to make a traditional beverage known as "tomi." This beverage is typically prepared with water, pulp, and sugar, and is consumed without prior treatment, making it a cost-effective, accessible, and ready-to-drink option (Konan et al., 2022; Ouattara et al., 2016; Yao et al., 2021). However, the consumption of this artisanal beverage could pose health risks if certain molds present in tamarind pulp are mycotoxin producers. To date, there have been very few studies focusing on the identification of molds in tamarind fruit, and none specifically address mycotoxin-producing fungi.
To address this gap, it is essential to identify the specific fungal species present in tamarind pulp and investigate the potential presence of mycotoxin-producing species. This will help to develop effective quality and food safety management strategies. Although sequencing is one of the most successful methods for identifying fungal species, it is prohibitively expensive when applied to large numbers of isolates. As a result, Restriction Fragment Length Polymorphisms (RFLP) analysis of the 5.8S region of the Internal Transcribed Spacer (ITS) region has emerged as the best alternative for grouping isolates, thereby reducing the number of strains requiring sequencing. In addition, this method has proven effective in identifying many fungal species and has been successfully extended to differentiate species within the same genus. Despite their low cost and relative ease of use, ITS-RFLP databases remain insufficient (Chanprasartsuk et al., 2013; Diguta et al., 2011, 2019).
This study aimed to characterize the toxigenic fungal flora of tamarind pulp to provide crucial information for developing improved management practices to ensure food safety and protect public health in the region.
Materials and methods
Sample collection
Tamarind samples were collected from wholesale markets in Abidjan, specifically from the Adjamé, Koumassi, and Abobo municipalities. From each of the three sampling sites, 60 tamarind samples (500 g each) were collected per wholesaler, resulting in a total of 180 samples. The samples were stored in coolers with ice packs at approximately 4 °C and transported to the laboratory for analysis.
pH measurement analysis
pH measurement was performed using the method described by Gacem (2011). A 20 g tamarind sample mixed with 40 ml distilled water was used to determine pH in 10 ml aliquots. Titratable Acidity (TA) was assessed according to ISO (1998) by titrating a 10 ml tamarind-water mixture with 1N sodium hydroxide (NaOH; S5881–500G, Sigma-Aldrich, USA) after adding phenolphthalein (P9750–25G, Sigma-Aldrich, USA). Moisture Content (MC) was determined by drying five g of tamarind at 105 °C for 24 h, following the method of Gacem (2011). Each experiment was carried out in triplicate.
Isolation of fungal and identification by conventional methods
Tamarind samples were inoculated by placing a seed directly onto Dichloran Rose Bengale Chloramphénicol (DRBC) agar (CM0727, Oxoid, UK) plates using sterile tweezers. Three Petri dishes per sample were prepared. The plates were incubated at 30 °C for 6 days, following the method described by Ghiasian et al. (2004). After incubation, characteristic mold colonies were purified by transferring a mycelium fragment from the colonies to the center of a new DRBC agar (CM0727, Oxoid, UK) plate. This plate was then incubated at 30 °C for an additional six days. Approximately 470 isolates were examined through macroscopic and microscopic observations, and they were stored at -20 °C.
DNA extraction and Polymerase Chain Reaction (PCR) reaction
For PCR assays, DNA extraction from 6-week-old mycelium on DRBC agar (CM0727, Oxoid, UK) was performed using the Dneasy PowerSoil® DNA isolation kit (12888-100, Qiagen, Germany), according to the manufacturer’s instructions. The PCR procedure targeting the 5.8S region of ITS region was conducted using the primers ITS1 (TCCGTAGGTGAACCTGCGG) and ITS4 (TCCTCCGCTTGATATGC). Each 50 µl PCR reaction contained 25 µl of one Taq 2X Master Mix (M0482S, New England Biolabs, UK), 0.5 µm of each primer, and 10-30 ng of fungal DNA. Amplification was carried out on a MasterCycler X50 (Eppendorf, Germany) with an initial denaturation at 95 °C for 5 min, followed by 30 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 40 s, and extension at 72 °C for 40 s. A final extension was performed at 72 °C for 5 min (Rao et al., 2020). The resulting PCR product was analyzed on a 1% agarose (A9539-100G, Sigma-Aldrich, USA) gel.
Taxonomic assignment of fungal isolates
PCR-amplified 5.8-ITS DNA fragments were sequenced using the primers ITS1 and ITS4. Eurofins Genomics (Germany) performed the sequencing of these fragments in both directions. The resulting nucleotide sequences have been submitted to the GenBank database, with accession numbers provided in Table 3. To identify the taxon, the sequences were compared with data from the National Center for Biotechnology Information (NCBI) database using the Basic Local Alignment Search Tool (BLAST) program (Altschul et al., 1990).
DNA digestion
PCR products were digested with endonucleases HaeIII (R6175, Promega, USA), as well as EcoRI (R0101S, New England Biolabs, UK), bslI (R0555S, New England Biolabs, UK), and Sau3AI (R0169S, New England Biolabs, UK) for 4 h at 37 °C. These enzymes were selected based on their ability to differentiate between isolates, as determined from the analysis of ITS sequences in the GenBank database. The resulting restriction fragments were analyzed using 2% agarose (A9539-100G, Sigma-Aldrich, USA) gels, and DNA bands were visualized under UV light. The sizes of the fragments were estimated by comparison with the Quick-Load Purple 50 bp DNA ladder (N0556S, New England Biolabs, UK).
Qualitative screening for toxin production in Aspergillus
-Ammonia vapor method
The Aspergillus isolates were grown on Potato Dextrose Agar (PDA) (CM0139, Oxoid, UK) and incubated at 28±2 °C for a period of 7 days, with two replicates for each sample. Following the incubation, the petri dishes were inverted, and 2 ml of concentrated ammonia solution (105422-1L, Sigma-Aldrich, USA) was added to the lid of each inverted plate. The plates were then exposed to ammonia vapour for 10-15 min. After this exposure, any changes in colony colour were observed and documented (Shekhar et al., 2017).
Aflatoxin gene screening
The genes aflD (Nor1-F: ACGGATCACTTAGCCAGCAC, Nor1-R: CTACCAGGGGAGTT GAGATCC), aflM (Ver1-F: CCGTTTAGATGGCAAAGTGG, Ver1-R: CTTTCAGGTGACC GAACGAT), aflO (OmtB(F)-F: GCCTTGACATGGAAACCATC, OmtB(F)-R: CCAAGATG GCCTGCTCTTTA), aflP (Omt1-F: GCCTTGCAAACACACTTTCA, Omt1-R: AGTTGTTG AACGCCCCAGT), aflQ (Ord1(P)-F: CGACTGTTGGCCTTTTCATT, Ord1(P)-R: ATAGCG AGGTTCCAGCGTAA), and aflR (AflR-F: CGAGTTGTGCCAGTTCAAAA, AflR-R: AAT CCTCGCCCACCATACTA) (Abd El-Aziz et al., 2021; Lin et al., 2013) were targeted for amplification in Aspergillus isolates identified as positive using the Ammonia vapor method. The PCR conditions followed the protocol established by Rao et al. (2020), consisting of an initial denaturation at 94 °C for 5 min, followed by 35 cycles of 30 s at 94 °C, 60 s at 55 °C, and 90 s at 72 °C, and concluded with a final extension at 72 °C for 7 min.
Data analysis
The data were analyzed using the Kruskal-Wallis test in R software (version 4.4.3; R Foundation for Statistical Computing) to assess differences between groups. Mean differences were evaluated, and statistical significance was determined at a significance level of p<0.05.
Results and discussion
The analysis revealed significant variations in pH, TA, and MC among the three districts (Table 1), reflecting the influence of local environmental conditions and processing methods on tamarind quality. Adjamé had a moderately acidic pH (2.2), higher than Koumassi (1.71) but lower than Abobo (2.40). These low pH values (below 3) indicate the acidic nature of tamarind attributed to the presence of organic acids, mainly tartaric acid, which imparts its characteristic sour taste (Hamacek et al., 2013; Obulesu and Bhattacharya, 2011; Soloviev et al., 2004). Our results are in agreement with those of other authors who also found that tamarind has an acidic pH (Asoiro et al., 2017; Devi and Boruah, 2020; Yao et al., 2021). The low pH can also be explained by the fact that, after picking, the tamarind pods undergo uncontrolled fermentation due to lactic acid bacteria. These produce organic acids that lower the pH of the tamarind (Bakayoko et al., 2024a, b). Koumassi had the highest TA (4.79%), followed by Adjamé (4.46%), and Abobo (4.03%). This range (4-5%) is typical of tamarind and highlights its acid profile, which can vary slightly depending on environmental and processing factors. In terms of MC, Abobo recorded the highest value (20.87%), while Adjamé (16.83%) and Koumassi (16.92%) had similar, lower values. This low MC could be explained by the fact that tamarind is a dried fruit (Asoiro et al., 2017; Devi and Boruah, 2020; Konan et al., 2022; Soloviev et al., 2004). Indeed, in Côte d'Ivoire, before tamarind is sold, the pods are dried in the sun (Aka et al., 2025). The low MC levels (14-20%) observed in all the samples are beneficial for shelf life, as they reduce enzymatic activity and microbial spoilage. This observation is consistent with previous findings, which indicate that dried or low-moisture foods (containing less than 25% MC) are more shelf-stable due to reduced water activity (Erkmen and Bozoglu, 2016).
Introduction
The tamarind tree Tamarindus indica, a monotypic species in the Fabaceae family, is native to tropical Africa. Its widespread cultivation in tropical regions (Diallo et al., 2010) highlights its importance across various sectors, including nutrition, traditional medicine, and cultural practices. In West Africa, tamarind plays a crucial role not only in traditional diets and therapeutic but also in the economic sustainability of local communities (Hamacek et al., 2013; Samarou et al., 2022a,b; Stege et al., 2011). Tamarind pulp is rich in macronutrients and micronutrients, particularly essential amino acids, and represents an excellent source of vitamin C. It is also rich in pectin and organic acids, particularly tartaric acid, and is extensively used in various food preparations such as sauces, jams, and fruit pastes (Bakayoko et al., 2024b; Devi and Boruah, 2020; Grollier et al., 1998; Hamacek et al., 2013; Samarou et al., 2022b). These culinary applications underscore not only the nutritional richness of tamarind pulp but also its versatility in local food traditions.
Despite its benefits, tamarind fruit faces significant challenges primarily due to its susceptibility to microbial contamination. Mold, prevalent in tropical environments, poses a substantial threat during the harvesting, storage, and processing stages of tamarind. This contamination not only affects the quality of the fruit but also poses serious food safety risks due to the potential production of mycotoxins. These secondary toxic metabolites produced by certain molds can lead to severe health issues including immune disorders, liver damage, and cancer (Kabak, 2009; Kouadio et al., 2014; Mafe and Büsselberg, 2024). Therefore, understanding the microbial flora associated with tamarind fruit, especially molds capable of producing mycotoxins, is crucial for ensuring food safety.
In Côte d'Ivoire, tamarind pulp, often more or less dried, is used to make a traditional beverage known as "tomi." This beverage is typically prepared with water, pulp, and sugar, and is consumed without prior treatment, making it a cost-effective, accessible, and ready-to-drink option (Konan et al., 2022; Ouattara et al., 2016; Yao et al., 2021). However, the consumption of this artisanal beverage could pose health risks if certain molds present in tamarind pulp are mycotoxin producers. To date, there have been very few studies focusing on the identification of molds in tamarind fruit, and none specifically address mycotoxin-producing fungi.
To address this gap, it is essential to identify the specific fungal species present in tamarind pulp and investigate the potential presence of mycotoxin-producing species. This will help to develop effective quality and food safety management strategies. Although sequencing is one of the most successful methods for identifying fungal species, it is prohibitively expensive when applied to large numbers of isolates. As a result, Restriction Fragment Length Polymorphisms (RFLP) analysis of the 5.8S region of the Internal Transcribed Spacer (ITS) region has emerged as the best alternative for grouping isolates, thereby reducing the number of strains requiring sequencing. In addition, this method has proven effective in identifying many fungal species and has been successfully extended to differentiate species within the same genus. Despite their low cost and relative ease of use, ITS-RFLP databases remain insufficient (Chanprasartsuk et al., 2013; Diguta et al., 2011, 2019).
This study aimed to characterize the toxigenic fungal flora of tamarind pulp to provide crucial information for developing improved management practices to ensure food safety and protect public health in the region.
Materials and methods
Sample collection
Tamarind samples were collected from wholesale markets in Abidjan, specifically from the Adjamé, Koumassi, and Abobo municipalities. From each of the three sampling sites, 60 tamarind samples (500 g each) were collected per wholesaler, resulting in a total of 180 samples. The samples were stored in coolers with ice packs at approximately 4 °C and transported to the laboratory for analysis.
pH measurement analysis
pH measurement was performed using the method described by Gacem (2011). A 20 g tamarind sample mixed with 40 ml distilled water was used to determine pH in 10 ml aliquots. Titratable Acidity (TA) was assessed according to ISO (1998) by titrating a 10 ml tamarind-water mixture with 1N sodium hydroxide (NaOH; S5881–500G, Sigma-Aldrich, USA) after adding phenolphthalein (P9750–25G, Sigma-Aldrich, USA). Moisture Content (MC) was determined by drying five g of tamarind at 105 °C for 24 h, following the method of Gacem (2011). Each experiment was carried out in triplicate.
Isolation of fungal and identification by conventional methods
Tamarind samples were inoculated by placing a seed directly onto Dichloran Rose Bengale Chloramphénicol (DRBC) agar (CM0727, Oxoid, UK) plates using sterile tweezers. Three Petri dishes per sample were prepared. The plates were incubated at 30 °C for 6 days, following the method described by Ghiasian et al. (2004). After incubation, characteristic mold colonies were purified by transferring a mycelium fragment from the colonies to the center of a new DRBC agar (CM0727, Oxoid, UK) plate. This plate was then incubated at 30 °C for an additional six days. Approximately 470 isolates were examined through macroscopic and microscopic observations, and they were stored at -20 °C.
DNA extraction and Polymerase Chain Reaction (PCR) reaction
For PCR assays, DNA extraction from 6-week-old mycelium on DRBC agar (CM0727, Oxoid, UK) was performed using the Dneasy PowerSoil® DNA isolation kit (12888-100, Qiagen, Germany), according to the manufacturer’s instructions. The PCR procedure targeting the 5.8S region of ITS region was conducted using the primers ITS1 (TCCGTAGGTGAACCTGCGG) and ITS4 (TCCTCCGCTTGATATGC). Each 50 µl PCR reaction contained 25 µl of one Taq 2X Master Mix (M0482S, New England Biolabs, UK), 0.5 µm of each primer, and 10-30 ng of fungal DNA. Amplification was carried out on a MasterCycler X50 (Eppendorf, Germany) with an initial denaturation at 95 °C for 5 min, followed by 30 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 40 s, and extension at 72 °C for 40 s. A final extension was performed at 72 °C for 5 min (Rao et al., 2020). The resulting PCR product was analyzed on a 1% agarose (A9539-100G, Sigma-Aldrich, USA) gel.
Taxonomic assignment of fungal isolates
PCR-amplified 5.8-ITS DNA fragments were sequenced using the primers ITS1 and ITS4. Eurofins Genomics (Germany) performed the sequencing of these fragments in both directions. The resulting nucleotide sequences have been submitted to the GenBank database, with accession numbers provided in Table 3. To identify the taxon, the sequences were compared with data from the National Center for Biotechnology Information (NCBI) database using the Basic Local Alignment Search Tool (BLAST) program (Altschul et al., 1990).
DNA digestion
PCR products were digested with endonucleases HaeIII (R6175, Promega, USA), as well as EcoRI (R0101S, New England Biolabs, UK), bslI (R0555S, New England Biolabs, UK), and Sau3AI (R0169S, New England Biolabs, UK) for 4 h at 37 °C. These enzymes were selected based on their ability to differentiate between isolates, as determined from the analysis of ITS sequences in the GenBank database. The resulting restriction fragments were analyzed using 2% agarose (A9539-100G, Sigma-Aldrich, USA) gels, and DNA bands were visualized under UV light. The sizes of the fragments were estimated by comparison with the Quick-Load Purple 50 bp DNA ladder (N0556S, New England Biolabs, UK).
Qualitative screening for toxin production in Aspergillus
-Ammonia vapor method
The Aspergillus isolates were grown on Potato Dextrose Agar (PDA) (CM0139, Oxoid, UK) and incubated at 28±2 °C for a period of 7 days, with two replicates for each sample. Following the incubation, the petri dishes were inverted, and 2 ml of concentrated ammonia solution (105422-1L, Sigma-Aldrich, USA) was added to the lid of each inverted plate. The plates were then exposed to ammonia vapour for 10-15 min. After this exposure, any changes in colony colour were observed and documented (Shekhar et al., 2017).
Aflatoxin gene screening
The genes aflD (Nor1-F: ACGGATCACTTAGCCAGCAC, Nor1-R: CTACCAGGGGAGTT GAGATCC), aflM (Ver1-F: CCGTTTAGATGGCAAAGTGG, Ver1-R: CTTTCAGGTGACC GAACGAT), aflO (OmtB(F)-F: GCCTTGACATGGAAACCATC, OmtB(F)-R: CCAAGATG GCCTGCTCTTTA), aflP (Omt1-F: GCCTTGCAAACACACTTTCA, Omt1-R: AGTTGTTG AACGCCCCAGT), aflQ (Ord1(P)-F: CGACTGTTGGCCTTTTCATT, Ord1(P)-R: ATAGCG AGGTTCCAGCGTAA), and aflR (AflR-F: CGAGTTGTGCCAGTTCAAAA, AflR-R: AAT CCTCGCCCACCATACTA) (Abd El-Aziz et al., 2021; Lin et al., 2013) were targeted for amplification in Aspergillus isolates identified as positive using the Ammonia vapor method. The PCR conditions followed the protocol established by Rao et al. (2020), consisting of an initial denaturation at 94 °C for 5 min, followed by 35 cycles of 30 s at 94 °C, 60 s at 55 °C, and 90 s at 72 °C, and concluded with a final extension at 72 °C for 7 min.
Data analysis
The data were analyzed using the Kruskal-Wallis test in R software (version 4.4.3; R Foundation for Statistical Computing) to assess differences between groups. Mean differences were evaluated, and statistical significance was determined at a significance level of p<0.05.
Results and discussion
The analysis revealed significant variations in pH, TA, and MC among the three districts (Table 1), reflecting the influence of local environmental conditions and processing methods on tamarind quality. Adjamé had a moderately acidic pH (2.2), higher than Koumassi (1.71) but lower than Abobo (2.40). These low pH values (below 3) indicate the acidic nature of tamarind attributed to the presence of organic acids, mainly tartaric acid, which imparts its characteristic sour taste (Hamacek et al., 2013; Obulesu and Bhattacharya, 2011; Soloviev et al., 2004). Our results are in agreement with those of other authors who also found that tamarind has an acidic pH (Asoiro et al., 2017; Devi and Boruah, 2020; Yao et al., 2021). The low pH can also be explained by the fact that, after picking, the tamarind pods undergo uncontrolled fermentation due to lactic acid bacteria. These produce organic acids that lower the pH of the tamarind (Bakayoko et al., 2024a, b). Koumassi had the highest TA (4.79%), followed by Adjamé (4.46%), and Abobo (4.03%). This range (4-5%) is typical of tamarind and highlights its acid profile, which can vary slightly depending on environmental and processing factors. In terms of MC, Abobo recorded the highest value (20.87%), while Adjamé (16.83%) and Koumassi (16.92%) had similar, lower values. This low MC could be explained by the fact that tamarind is a dried fruit (Asoiro et al., 2017; Devi and Boruah, 2020; Konan et al., 2022; Soloviev et al., 2004). Indeed, in Côte d'Ivoire, before tamarind is sold, the pods are dried in the sun (Aka et al., 2025). The low MC levels (14-20%) observed in all the samples are beneficial for shelf life, as they reduce enzymatic activity and microbial spoilage. This observation is consistent with previous findings, which indicate that dried or low-moisture foods (containing less than 25% MC) are more shelf-stable due to reduced water activity (Erkmen and Bozoglu, 2016).
Table 1: Physicochemical properties of tamarind pulps
| Districts | pH | TA (%) | MC (%) |
| Adjamé | 2.2±0,07 b | 4.46±0.02 b | 16.83±0.55 a |
| Koumassi | 1.71±0.05 a | 4.79±0.52 c | 16.92±0.53 a |
| Abobo | 2.40±0.06 c | 4.03±0.10 a | 20.87±0.54 b |
| In a column, mean± Standard Deviation (SD) values followed by different superscript letters are significantly different according to the Kruskal-Wallis test (p<0.05). MC=Moisture Content; TA=Titratable Acidity |
|||
PCR amplification of the 5.8S region of the ITS using primers ITS1 and ITS4, produced a 600-bp band (Figure 1) for all 470 fungal isolates. Based on macroscopic and microscopic characteristics, as well as RFLP profiles, five groups were identified. Group I w::as char::acterized by fluffy yellow colonies with a white margin and a colorless reverse. Microscopically, it exhibited septate, branched mycelia, and biseriate conidial heads with globose vesicles (Table 2).
Table 2: Macroscopic and microscopic appearances of tamarind fungal isolates
| Group | Macroscopic appearance | Microscopic appearance |
| Group I | Fluffy colonies, yellow with white margin, colorless reverse | Septate, branched mycelia; biseriate, radiate conidial heads; globose vesicles |
| Group II | Powdery green colonies with white margin, pale pink reverse, medium forms | Colorless, septate, branched mycelia; biseriate conidial heads with spherical vesicles |
| Group III | Flat gray-yellow colonies with colorless reverse, small forms | Colorless, septate mycelia; brush-like ramified conidial heads |
| Group IV | Granular black colonies with white margin, beige reverse, large forms | Septate, branched mycelia; fast-growing thallus; globose, brown conidia; vesicles sometimes slightly flattened |
| Group V | Powdery brown colonies spreading on medium, beige reverse | Septate, branched mycelia; globose conidiophores |

Figure 1: Polymerase Chain Reaction (PCR) product of the Internal Transcribed Spacer (ITS-5.8S) rDNA region. L: 100 bp DNA ladder; lanes 1-10: PCR products from 10 out of 470 fungal isolates; lane 11: water control. The gel shows a 600 bp PCR product corresponding to the ITS-5.8S rDNA region of the fungal isolates
Enzymatic digestion using HaeIII, EcoRI, Sau3AI, and BslI produced fragments of 350 and 80 bp, 310 and 280 bp, 320 and 250 bp, and 435 and 90 bp, respectively (Table 3). BLAST analysis of the 5.8S-ITS sequence identified the species as Aspergillus melleus (99.12-100% identity) from the subgenus Circumdati and section Circumdati, with a prevalence of 20%. The identification of A. melleus highlights its potential role as a tamarind contaminant. While no pathogenic potential is commonly attributed to this species, its adaptation to low-moisture environments may explain its presence in tamarind samples. Group II exhibited powdery green colonies with a white margin and pale pink reverse. Microscopically, it displayed colorless, septate, branched mycelium, and biseriate conidial heads with spherical vesicles. Enzymatic digestion revealed fragments of 260, 110, and 70 bp for HaeIII; 295 and 280 bp for EcoRI; 330 and 180 bp for Sau3AI; and 350 and 160 bp for BslI. This group was identified as Monascus pilosus (99.65-100% identity), a species from the genus Monascus within the family Trichocomaceae, with a prevalence of 15%. Although M. pilosus is not commonly associated with mycotoxin production, its presence highlights the diversity of fungal species capable of contaminating tamarind. Group III showed flat gray-yellow colonies with colorless reverse and small forms. Microscopically, it exhibited colorless, septate mycelium, and brush-like ramified conidial heads. Fragments of 290, 100, and 70 bp for HaeIII; 300 and 290 bp for EcoRI; 230, 150, and 90 bp for Sau3AI; and 380 and 180 bp for BslI were observed. BLAST analysis confirmed the species as Aspergillus aflatoxiformans (100% identity) (Table 3) from the section Flavi, with a prevalence of 5%. A. aflatoxiformans has been identified as a producer of aflatoxins B1, B2, G1, and G2 and is commonly detected in foods across Africa (Frisvad et al., 2019). Among the major mycotoxins, aflatoxins, classified as furanocoumarin-derived polyketides, are considered to be the most toxic and carcinogenic. Aflatoxins are commonly found contaminating various agricultural products and present significant health risks to both humans and livestock (Navale et al., 2021). Group IV showed granular black colonies with a white margin and beige reverse. It featured septate, branched mycelium, a fast-growing thallus, globose brown conidia, and slightly flattened vesicles. The restriction fragments obtained were 290, 90 and 70 bp for HaeIII; 300 and 290 bp for EcoRI; 230, 210 and 90 bp for Sau3AI; and 300, 180, and 70 bp for BslI. The species was identified as Aspergillus niger (99.62-100% identity) from the section Nigri, with a prevalence of 20%. A. niger is known for its ability to produce ochratoxin A (OTA), on natural substrates, although species within the Nigri section generally produce lower levels of this toxin. OTA is commonly formed during storage of crops and has been identified as neurotoxic, hepatotoxic, immunotoxic, genotoxic, embryotoxic, teratogenic, and carcinogenic in both humans and animals (Navale et al., 2021). Group V displayed powdery brown colonies spreading on the medium with a beige reverse. Microscopically, it featured septate, branched mycelium, and globose conidiophores. This group exhibited restriction patterns identical to Group IV. The species was identified as Aspergillus costaricensis (99.67–100% identity) from the section Nigri, with a prevalence of 40%.
A recent study by Bian et al. (2022) reclassified 14 Aspergillus species within the Nigri section into six species, with A. costaricensis being reclassified as Aspergillus tubingensis. The work of Kizis et al. (2014) showed that the polymorphism between A. niger and A. tubingensis could be distinguished using the enzyme RsaI. Therefore, A. niger and A. costaricensis could potentially be distinguished in our study using the RsaI enzyme. However, it should be noted that no previous study has documented the RFLP profiles of the fungi analyzed here. Therefore, this study contributes to the enrichment of existing RFLP profile databases for fungi and underscores the utility of combining morphological identification, RFLP profiling, and sequencing for fungal species characterization. While RFLP is cost-effective for reducing sequencing needs, its limitations in distinguishing closely related species, such as A. niger and A. costaricensis, were evident. The prominence of Aspergillus species across tamarind samples can be attributed to their ability to thrive in low-moisture environments, as observed in grains where moisture levels below 13% did not significantly inhibit fungal growth (Nji et al., 2022). The detection of mycotoxin-producing species, such as A. niger and A. aflatoxiformans, highlights potential health risks and underscores the importance of storage and quality control measures for tamarind products.
Table 3: Fungal identities and restriction profiles
| Group | Restriction enzymes | Identity obtained after sequencing |
% Identity |
Prevalence (%) |
GenBank accession number |
|||
| HaeIII | EcoRI | Sau3AI | BslI | |||||
| Group I | 350+80 | 310+280 | 320+250 | 435+90 | Aspergillus melleus | 99.12-100 | 13.77 | PQ512171 |
| Group II | 260+110+70 | 295+280 | 330+180 | 350+160 | Monascus pilosus | 99.65-100 | 11.33 | PQ512170 |
| Group III | 290+100+70 | 300+290 | 230+150+90 | 380+180 | Aspergillus aflatoxiformans | 100 | 3.60 | PQ512167 |
| Group IV | 290+90+70 | 300+290 | 230+210+90 | 300+180+70 | Aspergillus costaricensis | 99.67-100 | 45.65 | PQ512168 |
| Group V | 290+90+70 | 300+290 | 230+210+90 | 300+180+70 | Aspergillus niger | 99.62-100 | 25.65 | PQ512169 |
Cultures of Aspergillus from the section Flavi were exposed to ammonia vapor. The appearance of a plum-red coloration on the underside of Petri dishes after exposure to ammonium hydroxide is an indicator of aflatoxigenic potential. This color change was observed in A. aflatoxiformans. The color change is attributed to anthraquinone intermediates involved in aflatoxin biosynthesis, which react under alkaline conditions to produce a red hue. In contrast, non-aflatoxigenic isolates do not exhibit any color change, confirming their inability to produce aflatoxins (Ronoh et al., 2024). The observed variations in color intensity are likely linked to differences in toxin production levels among the isolates. The plum-red coloration observed in this study may indicate lower aflatoxin production levels. However, this qualitative method does not determine the specific type of aflatoxin produced by A. aflatoxiformans. The color of the cultures changed from beige to plum red following exposure (Figure 2).

Figure 2: Qualitative screening of aflatoxin production in Aspergillus aflatoxiformans plates exposed to ammonia vapor (Left: beige control, Right: color change to plum red in isolate)
To confirm the toxinogenic potential of these isolates, six key aflatoxin pathway genes were screened. The results revealed the presence of aflD, aflM, aflO, aflP, aflQ, and aflR gene (Figure 3) in our A. aflatoxiformans isolates. These genes are known to play a critical role in regulating aflatoxin biosynthesis in fungal species (Abd El-Aziz et al., 2021; Musangi et al., 2024). The amplification of these genes in the isolates confirms their potential to produce aflatoxins. These specific genes were selected as targets because of their fundamental involvement in the aflatoxin biosynthetic pathway. The aflR gene is an important regulator of the aflatoxin biosynthesis gene cluster. The presence of the aflR gene is still of concern because a presumably atoxigenic fungus may later acquire the ability to produce aflatoxins. In addition, aflatoxin production is known to be influenced by other factors such as pH, substrate, temperature, oxidative stress, and water availability. The aflO gene encodes an O-methyltransferase enzyme that converts demethylsterigmatocystin (DMST) to sterigmatocystin (ST) and dihydrodemethylsterigmatocystin (DHDMST) to dihydrosterigmatocystin (DHST) (Ronoh et al., 2024). Each gene in the aflatoxin biosynthetic pathway has a specific function: aflD, aflF, and aflM encode enzymes involved in early steps of toxin synthesis, while aflO, aflP, and aflQ are involved in intermediate and final conversions. The aflR gene acts as a key transcriptional regulator, coordinating the expression of the entire gene cluster (Abdel-Hadi et al., 2011).

Figure 3: Polymerase Chain Reaction (PCR) product targeting the aflR gene in Aspergillus aflatoxiformans. L: 50 bp DNA Ladder; Lane 1: aflD gene; Lane 2: aflQ gene; Lane 3: aflM gene; Lane 4: aflP gene; Lane 5: aflR gene; Lane 6: aflO gene
Conclusions
The results of this study indicate that tamarind is a highly acidic fruit with a relatively low MC, making it well-suited for prolonged storage. The phenotypic identification of fungal colonies identified five species, namely M. pilosus, A. melleus, A. aflatoxiformans, A. niger, and A. costaricensis. Whereas, molecular identification yielded four distinct fungal profiles, highlighting that RFLP is helpful for grouping isolates. However, closely related species such as A. niger and A. costaricensis were difficult to distinguish. Contamination by Aspergillus species, particularly those capable of producing mycotoxins, raises food safety concerns, especially under suboptimal storage conditions. Future research should focus on monitoring mycotoxin levels in tamarind products and implementing better storage practices to reduce fungal contamination and ensure consumer safety.
Author contributions
B.B., T.L.S.A., and S.A.G. designed the study; B.B. acquired funding, conducted data curation and analyzed the data; B.B., T.L.S.A., K.M.C.N., M.K., and L.I.A.T. conducted the experimental work, and wrote the original draft; T.L.S.A. and S.A.G. supervised the research and validated the results; S.A.G. and M.K.D. reviewed and edited the manuscript. All authors read and approved the final manuscript.
Acknowledgments
The authors would like to express their sincere gratitude to the traders at the Abidjan wholesale markets from whom the tamarind was collected.
Conflicts of Interest
The authors declare that there is no conflict of interest.
Funding
This study did not receive any specific grant from funding agencies in the public, commercial, or non-profit sectors.
Ethical consideration
Not applicable.
References
Abd El-Aziz A.R.M., Shehata S.M., Hisham S.M., Alobathani A.A. (2021). Molecular profile of aflatoxigenic and non-aflatoxigenic isolates of Aspergillus flavus isolated from stored maize. Saudi Journal of Biological Sciences. 28: 1383–1391. [DOI: 10.1016/j.sjbs.2020.11.073]
Abdel-Hadi A., Schmidt-Heydt M., Parra R., Geisen R., Magan N. (2011). A systems approach to model the relationship between aflatoxin gene cluster expression, environmental factors, growth and toxin production by Aspergillus flavus. Journal of The Royal Society Interface. 9: 757–767. [DOI: 10.1098/rsif.2011.0482]
Aka S., Yao K.F., Amoikon L.-S.T., Balla B., Bouatenin J.-P. (2025). Socio-economic, physico-chemical and microbiological study of tamarind pulps (Tamarindus indica) sold in Adjamé and Boundiali (Côte d’Ivoire). Moroccan Journal of Agricultural and Veterinary Sciences. 13: 19-26. [DOI: 10.5281/zenodo. 15005651]
Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. (1990). Basic local alignment search tool. Journal of Molecular Biology. 215: 403-410. [DOI: 10.1016/S0022-2836(05)80360-2]. available at http://blast.ncbi.nlm.nih.gov/Blast.cgi.
Asoiro F.U., Ezeoha S.L., Ezenne G.I., Ugwu C.B. (2017). Chemical and mechanical properties of velvet tamarind fruit (Dialium guineense). Nigerian Journal of Technology. 36: 252-260. [DOI: 10.4314/njt.v36i1.30]
Bakayoko N.L., Soumahoro S., Tiekoura K.B., Ouattara M.B., Guede K.B., Coulibaly A.M., Guessennd N.K., Camara-Koussemon C (2024a). Diversity of lactic acid bacteria involved in the fermentation of the tamarind fruit (Tamarindus indica) Côte d’Ivoire. International Journal of Advanced Research. 12: 766-774. [DOI:10.21474/IJAR01/18938]
Bakayoko N.L., Tiekoura K.B., Soumahoro S., Toure A., Guessennd N.K., Camara-Koussemon C (2024b). Food and therapeutic uses of tamarind fruit in Korhogo and Abidjan, Côte d'Ivoire. Afrique Science. 24: 47-59.
Bian C., Kusuya Y., Sklenář F., D’hooge E., Yaguchi T., Ban S., Visagie C.M., Houbraken J., Takahashi H., Hubka V. (2022). Reducing the number of accepted species in Aspergillus series Nigri. Studies in Mycology. 102: 95-132. [DOI: 10.3114/sim.2022.102.03]
Chanprasartsuk O., Prakitchaiwattana C., Sanguandeekul R. (2013). Comparison of methods for identification of yeasts isolated during spontaneous fermentation of freshly crushed pineapple juices. Journal of Agricultural Science and Technology. 15: 1779-1790. URL: http://jast.modares.ac.ir/article-23-5776-en.html.
Devi B., Boruah T. (2020). Tamarind (Tamarindus indica). In: Nayik G.A., Gull A. (Editors). Antioxidants in fruits: properties and health benefits. Springer Nature, Singapore. pp: 317-332. [DOI: 10.1007/978-981-15-7285-2_16]
Diallo B.O., Joly H.I., Key D.M., Hossaert-Mckey M., Chevallier M.H. (2010). Change in biometric characters of seeds and seedlings of nine provenances of Tamarindus indica L. (Caesalpinioideae). Fruits. 65: 153-167. [DOI: 10.1051/fruits/2010010].
Diguță C.F., Proca I.G., Jurcoane S. Matei F. (2019). Molecular characterization by PCR-RFLP of indigenous fungal isolates from hypersaline stream water in România. Folia Microbiological. 64: 407-414. [DOI: 10.1007/s12223-018-0664-6]
Diguta C.F., Vincent B., Guilloux-Benatier M., Alexandre H., Rousseaux S. (2011). PCR ITS-RFLP: a useful method for identifying filamentous fungi isolates on grapes. Food Microbiology. 28: 1145-1154. [DOI:10.1016/j.fm.2011.03.006]
Erkmen O., Bozoglu T. F. (2016). Food preservation by reducing water activity. In: Erkmen O., Bozoglu T.F. (Editors). Food microbiology: principles into practice. John Wiley and Sons, Chichester, West Sussex. pp: 44-58. [DOI: 10.1002/9781119237860.ch30]
Frisvad J.C., Hubka V., Ezekiel C.N., Hong S.-B., Nováková A., Chen A.J., Arzanlou M., Larsen T.O., Sklenář F., Mahakarnchanakul W., Samson R.A., Houbraken J. (2019). Taxonomy of Aspergillus section Flavi and their production of aflatoxins, ochratoxins and other mycotoxins. Studies in Mycology. 93: 1-63. [DOI: 10.1016/j.simyco.2018.06.001]
Gacem M. (2011). Contribution to study of the antifungal and antimycotoxigenic activity of methanol and aqueous extract of seeds of Citrullus colocynthis against some fungi isolated from wheat stored. Doctoral dissertation. Kasdi Merbah University of Ouargla, Algeria. 149. URL: https://dspace.univ-ouargla.dz/ jspui/bitstream/123456789/487/1/GACEM_Mohamed_Amine.pdf.
Ghiasian S.A., Kord-Bacheh P., Rezayat S.M., Maghsood A.H., Taherkhani H. (2004). Mycoflora of Iranian maize harvested in the main production areas in 2000. Mycopathologia. 158: 113-121. [DOI: 10.1023/B:MYCO.0000038425.95049.03]
Grollier C., Debien C., Dornier M., Reynes M. (1998). Prominent characteristics and possible uses of the tamarind. Fruits. 53: 271-280.
Hamacek F.R., Santos P.R.G., Cardoso L.D.M., Pinheiro-Sant’Ana H.M. (2013). Nutritional composition of tamarind (Tamarindus indica L.) from the Cerrado of Minas Gerais, Brazil. Fruits. 68: 381-395. [DOI: 10.1051/fruits/2013083]
International Organization for Standardization (ISO). (1998). Fruit and vegetable products – determination of titratable acidity (ISO 750:1998). URL: https://cdn.standards.iteh.ai/samples/22569/eef01c3051f247398eb411358ccf25d1/ISO-750-1998.pdf.
Kabak B. (2009). The fate of mycotoxins during thermal food processing. Journal of the Science of Food and Agriculture. 89: 549-554. [DOI: 10.1002/jsfa.3491]
Kizis D., Natskoulis P., Nychas G.-J.E., Panagou E.Z. (2014). Biodiversity and ITS-RFLP characterisation of Aspergillus section Nigri isolates in grapes from four traditional grape-producing areas in Greece. PLOS ONE. 9: e93923. [DOI: 10.1371/journal.pone.0093923]
Konan G.A.J., Nimaga D., Kouakou K.N., N’dri Y.D., Amani N.G. (2022). Identification of the main dishes made from the pulp of tomi (Tamarindus indica L) consumed in the Savannah Region of Côte d'Ivoire. Journal of Experimental Agriculture International. 44: 224-237. [DOI: 10.9734/JEAI/2022/v44i1030899]
Kouadio J.H., Lattanzio V.M.T., Ouattara D., Kouakou B., Visconti A. (2014). Assessment of mycotoxin exposure in Côte d’Ivoire (Ivory Coast) through multi-biomarker analysis and possible correlation with food consumption patterns. Toxicology International. 21: 248-257. [DOI: 10.4103/0971-6580.155336]
Lin J.-Q., Zhao X.-X., Wang C.-C., Xie Y., Li G.-H., He Z.-M. (2013). 5-Azacytidine inhibits aflatoxin biosynthesis in Aspergillus flavus. Annals of Microbiology. 63: 763–769. [DOI: 10.1007/s13213-012-0531-7]
Mafe A.N., Büsselberg D. (2024). Mycotoxins in food: cancer risks and strategies for control. Foods. 13: 3502. [DOI: 10.3390/foods13213502]
Musangi C.R., Juma B.S., Mukhebi D.W., Isoe E.M., Kibiti C.M., Mbinda W.M. (2024). Aspergillus population diversity and its role in aflatoxin contamination of cashew nuts from coastal Kenya. PLOS ONE. 19: e0292519. [DOI: 10.1371/journal.pone.0292519]
Navale V., Vamkudoth K.R., Ajmera S., Dhuri V. (2021). Aspergillus derived mycotoxins in food and the environment: prevalence, detection, and toxicity. Toxicology Reports. 8: 1008–1030. [DOI: 10.1016/j.toxrep.2021.04.013]
Nji N.Q., Christianah A.M., Njie A.C., Mulunda M. (2022). Biodiversity and distribution of Aspergillus and their toxins in maize from western and eastern regions of South Africa. Advances in Microbiology. 12: 121-149. [DOI : 10.4236/aim.2022.123011]
Obulesu M., Bhattacharya S. (2011). Color changes of tamarind (Tamarindus indica L.) pulp during fruit development, ripening, and storage. International Journal of Food Properties. 14: 538-549. [DOI: 10.1080/10942910903262129].
Ouattara N.D., Gaille E., Stauffer F.W., Bakayoko A. (2016). Floristic diversity and ethnobotany of edible wild plants in the Department of Bondoukou (North-Eastern Côte d'Ivoire). Journal of Applied Biosciences. 98: 9284–9300. [DOI: 10.4314/jab.v98i1.5]
Rao K.R., Vipin A.V., Venkateswaran G. (2020). Molecular profile of non-aflatoxigenic phenotype in native strains of Aspergillus flavus. Archives of Microbiology. 202: 1143–1155. [DOI: 10.1007/s00203-020-01822-1]
Ronoh P.K., Toroitich F.J., Makonde H.M., Lelmen E.K., Obonyo M.A. (2024). Reliability of the chemical, metabolic, and molecular methods in discriminating aflatoxigenic from non-aflatoxigenic Aspergillus isolates. The Microbe. 4: 100115. [DOI: 10.1016/j.microb.2024.100115]
Samarou M., Atakpama W., Atato A., Pessinaba Mamoudou M., Batawila K., Akpagana K. (2022a). Socio-economic value of tamarind (Tamarindus indica) in ecological zone I of Togo. Moroccan Journal of Agricultural and Veterinary Sciences. 10 : 272-281.
Samarou M., Atakpama W., Batawila K., Akpagana K. (2022b). State of knowledge on Tamarindus indica L. (Fabaceae). Agronomie Africaine SP. 34: 313–323.
Shekhar M., Singh N., Dutta R., Kumar S., Mahajan V. (2017). Comparative study of qualitative and quantitative methods to determine toxicity level of Aspergillus flavus isolates in maize. PLOS ONE. 12: e0189760. [DOI: 10.1371/journal.pone. 0189760]
Soloviev P., Niang T.D., Gaye A., Totte A. (2004). Variability of fruit physicochemical characters for three harvested woody species in Senegal: Adansonia digitata, Balanites aegyptiaca and Tamarindus indica. Fruits. 59: 109-119. [DOI : 10.1051/fruits:2004011]
Stege C.V.D., Prehsler S., Hartl A., Vogl C.R. (2011). Tamarind (Tamarindus indica L.) in the traditional West African diet: not just a famine food. Fruits. 66: 171–185. [DOI: 10.1051/ fruits/2011025]
Yao K.M., Kambire O., Coulibaly K.R., Diomande A., Koffi-nevry R. (2021). Microbiological and physicochemical quality of «Tomi»: an artisanal drink made from Tamarindus indica pulp, sold in Korhogo (Côte d’Ivoire). International Journal of Innovation and Applied Studies. 34: 551-560.
The results of this study indicate that tamarind is a highly acidic fruit with a relatively low MC, making it well-suited for prolonged storage. The phenotypic identification of fungal colonies identified five species, namely M. pilosus, A. melleus, A. aflatoxiformans, A. niger, and A. costaricensis. Whereas, molecular identification yielded four distinct fungal profiles, highlighting that RFLP is helpful for grouping isolates. However, closely related species such as A. niger and A. costaricensis were difficult to distinguish. Contamination by Aspergillus species, particularly those capable of producing mycotoxins, raises food safety concerns, especially under suboptimal storage conditions. Future research should focus on monitoring mycotoxin levels in tamarind products and implementing better storage practices to reduce fungal contamination and ensure consumer safety.
Author contributions
B.B., T.L.S.A., and S.A.G. designed the study; B.B. acquired funding, conducted data curation and analyzed the data; B.B., T.L.S.A., K.M.C.N., M.K., and L.I.A.T. conducted the experimental work, and wrote the original draft; T.L.S.A. and S.A.G. supervised the research and validated the results; S.A.G. and M.K.D. reviewed and edited the manuscript. All authors read and approved the final manuscript.
Acknowledgments
The authors would like to express their sincere gratitude to the traders at the Abidjan wholesale markets from whom the tamarind was collected.
Conflicts of Interest
The authors declare that there is no conflict of interest.
Funding
This study did not receive any specific grant from funding agencies in the public, commercial, or non-profit sectors.
Ethical consideration
Not applicable.
References
Abd El-Aziz A.R.M., Shehata S.M., Hisham S.M., Alobathani A.A. (2021). Molecular profile of aflatoxigenic and non-aflatoxigenic isolates of Aspergillus flavus isolated from stored maize. Saudi Journal of Biological Sciences. 28: 1383–1391. [DOI: 10.1016/j.sjbs.2020.11.073]
Abdel-Hadi A., Schmidt-Heydt M., Parra R., Geisen R., Magan N. (2011). A systems approach to model the relationship between aflatoxin gene cluster expression, environmental factors, growth and toxin production by Aspergillus flavus. Journal of The Royal Society Interface. 9: 757–767. [DOI: 10.1098/rsif.2011.0482]
Aka S., Yao K.F., Amoikon L.-S.T., Balla B., Bouatenin J.-P. (2025). Socio-economic, physico-chemical and microbiological study of tamarind pulps (Tamarindus indica) sold in Adjamé and Boundiali (Côte d’Ivoire). Moroccan Journal of Agricultural and Veterinary Sciences. 13: 19-26. [DOI: 10.5281/zenodo. 15005651]
Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. (1990). Basic local alignment search tool. Journal of Molecular Biology. 215: 403-410. [DOI: 10.1016/S0022-2836(05)80360-2]. available at http://blast.ncbi.nlm.nih.gov/Blast.cgi.
Asoiro F.U., Ezeoha S.L., Ezenne G.I., Ugwu C.B. (2017). Chemical and mechanical properties of velvet tamarind fruit (Dialium guineense). Nigerian Journal of Technology. 36: 252-260. [DOI: 10.4314/njt.v36i1.30]
Bakayoko N.L., Soumahoro S., Tiekoura K.B., Ouattara M.B., Guede K.B., Coulibaly A.M., Guessennd N.K., Camara-Koussemon C (2024a). Diversity of lactic acid bacteria involved in the fermentation of the tamarind fruit (Tamarindus indica) Côte d’Ivoire. International Journal of Advanced Research. 12: 766-774. [DOI:10.21474/IJAR01/18938]
Bakayoko N.L., Tiekoura K.B., Soumahoro S., Toure A., Guessennd N.K., Camara-Koussemon C (2024b). Food and therapeutic uses of tamarind fruit in Korhogo and Abidjan, Côte d'Ivoire. Afrique Science. 24: 47-59.
Bian C., Kusuya Y., Sklenář F., D’hooge E., Yaguchi T., Ban S., Visagie C.M., Houbraken J., Takahashi H., Hubka V. (2022). Reducing the number of accepted species in Aspergillus series Nigri. Studies in Mycology. 102: 95-132. [DOI: 10.3114/sim.2022.102.03]
Chanprasartsuk O., Prakitchaiwattana C., Sanguandeekul R. (2013). Comparison of methods for identification of yeasts isolated during spontaneous fermentation of freshly crushed pineapple juices. Journal of Agricultural Science and Technology. 15: 1779-1790. URL: http://jast.modares.ac.ir/article-23-5776-en.html.
Devi B., Boruah T. (2020). Tamarind (Tamarindus indica). In: Nayik G.A., Gull A. (Editors). Antioxidants in fruits: properties and health benefits. Springer Nature, Singapore. pp: 317-332. [DOI: 10.1007/978-981-15-7285-2_16]
Diallo B.O., Joly H.I., Key D.M., Hossaert-Mckey M., Chevallier M.H. (2010). Change in biometric characters of seeds and seedlings of nine provenances of Tamarindus indica L. (Caesalpinioideae). Fruits. 65: 153-167. [DOI: 10.1051/fruits/2010010].
Diguță C.F., Proca I.G., Jurcoane S. Matei F. (2019). Molecular characterization by PCR-RFLP of indigenous fungal isolates from hypersaline stream water in România. Folia Microbiological. 64: 407-414. [DOI: 10.1007/s12223-018-0664-6]
Diguta C.F., Vincent B., Guilloux-Benatier M., Alexandre H., Rousseaux S. (2011). PCR ITS-RFLP: a useful method for identifying filamentous fungi isolates on grapes. Food Microbiology. 28: 1145-1154. [DOI:10.1016/j.fm.2011.03.006]
Erkmen O., Bozoglu T. F. (2016). Food preservation by reducing water activity. In: Erkmen O., Bozoglu T.F. (Editors). Food microbiology: principles into practice. John Wiley and Sons, Chichester, West Sussex. pp: 44-58. [DOI: 10.1002/9781119237860.ch30]
Frisvad J.C., Hubka V., Ezekiel C.N., Hong S.-B., Nováková A., Chen A.J., Arzanlou M., Larsen T.O., Sklenář F., Mahakarnchanakul W., Samson R.A., Houbraken J. (2019). Taxonomy of Aspergillus section Flavi and their production of aflatoxins, ochratoxins and other mycotoxins. Studies in Mycology. 93: 1-63. [DOI: 10.1016/j.simyco.2018.06.001]
Gacem M. (2011). Contribution to study of the antifungal and antimycotoxigenic activity of methanol and aqueous extract of seeds of Citrullus colocynthis against some fungi isolated from wheat stored. Doctoral dissertation. Kasdi Merbah University of Ouargla, Algeria. 149. URL: https://dspace.univ-ouargla.dz/ jspui/bitstream/123456789/487/1/GACEM_Mohamed_Amine.pdf.
Ghiasian S.A., Kord-Bacheh P., Rezayat S.M., Maghsood A.H., Taherkhani H. (2004). Mycoflora of Iranian maize harvested in the main production areas in 2000. Mycopathologia. 158: 113-121. [DOI: 10.1023/B:MYCO.0000038425.95049.03]
Grollier C., Debien C., Dornier M., Reynes M. (1998). Prominent characteristics and possible uses of the tamarind. Fruits. 53: 271-280.
Hamacek F.R., Santos P.R.G., Cardoso L.D.M., Pinheiro-Sant’Ana H.M. (2013). Nutritional composition of tamarind (Tamarindus indica L.) from the Cerrado of Minas Gerais, Brazil. Fruits. 68: 381-395. [DOI: 10.1051/fruits/2013083]
International Organization for Standardization (ISO). (1998). Fruit and vegetable products – determination of titratable acidity (ISO 750:1998). URL: https://cdn.standards.iteh.ai/samples/22569/eef01c3051f247398eb411358ccf25d1/ISO-750-1998.pdf.
Kabak B. (2009). The fate of mycotoxins during thermal food processing. Journal of the Science of Food and Agriculture. 89: 549-554. [DOI: 10.1002/jsfa.3491]
Kizis D., Natskoulis P., Nychas G.-J.E., Panagou E.Z. (2014). Biodiversity and ITS-RFLP characterisation of Aspergillus section Nigri isolates in grapes from four traditional grape-producing areas in Greece. PLOS ONE. 9: e93923. [DOI: 10.1371/journal.pone.0093923]
Konan G.A.J., Nimaga D., Kouakou K.N., N’dri Y.D., Amani N.G. (2022). Identification of the main dishes made from the pulp of tomi (Tamarindus indica L) consumed in the Savannah Region of Côte d'Ivoire. Journal of Experimental Agriculture International. 44: 224-237. [DOI: 10.9734/JEAI/2022/v44i1030899]
Kouadio J.H., Lattanzio V.M.T., Ouattara D., Kouakou B., Visconti A. (2014). Assessment of mycotoxin exposure in Côte d’Ivoire (Ivory Coast) through multi-biomarker analysis and possible correlation with food consumption patterns. Toxicology International. 21: 248-257. [DOI: 10.4103/0971-6580.155336]
Lin J.-Q., Zhao X.-X., Wang C.-C., Xie Y., Li G.-H., He Z.-M. (2013). 5-Azacytidine inhibits aflatoxin biosynthesis in Aspergillus flavus. Annals of Microbiology. 63: 763–769. [DOI: 10.1007/s13213-012-0531-7]
Mafe A.N., Büsselberg D. (2024). Mycotoxins in food: cancer risks and strategies for control. Foods. 13: 3502. [DOI: 10.3390/foods13213502]
Musangi C.R., Juma B.S., Mukhebi D.W., Isoe E.M., Kibiti C.M., Mbinda W.M. (2024). Aspergillus population diversity and its role in aflatoxin contamination of cashew nuts from coastal Kenya. PLOS ONE. 19: e0292519. [DOI: 10.1371/journal.pone.0292519]
Navale V., Vamkudoth K.R., Ajmera S., Dhuri V. (2021). Aspergillus derived mycotoxins in food and the environment: prevalence, detection, and toxicity. Toxicology Reports. 8: 1008–1030. [DOI: 10.1016/j.toxrep.2021.04.013]
Nji N.Q., Christianah A.M., Njie A.C., Mulunda M. (2022). Biodiversity and distribution of Aspergillus and their toxins in maize from western and eastern regions of South Africa. Advances in Microbiology. 12: 121-149. [DOI : 10.4236/aim.2022.123011]
Obulesu M., Bhattacharya S. (2011). Color changes of tamarind (Tamarindus indica L.) pulp during fruit development, ripening, and storage. International Journal of Food Properties. 14: 538-549. [DOI: 10.1080/10942910903262129].
Ouattara N.D., Gaille E., Stauffer F.W., Bakayoko A. (2016). Floristic diversity and ethnobotany of edible wild plants in the Department of Bondoukou (North-Eastern Côte d'Ivoire). Journal of Applied Biosciences. 98: 9284–9300. [DOI: 10.4314/jab.v98i1.5]
Rao K.R., Vipin A.V., Venkateswaran G. (2020). Molecular profile of non-aflatoxigenic phenotype in native strains of Aspergillus flavus. Archives of Microbiology. 202: 1143–1155. [DOI: 10.1007/s00203-020-01822-1]
Ronoh P.K., Toroitich F.J., Makonde H.M., Lelmen E.K., Obonyo M.A. (2024). Reliability of the chemical, metabolic, and molecular methods in discriminating aflatoxigenic from non-aflatoxigenic Aspergillus isolates. The Microbe. 4: 100115. [DOI: 10.1016/j.microb.2024.100115]
Samarou M., Atakpama W., Atato A., Pessinaba Mamoudou M., Batawila K., Akpagana K. (2022a). Socio-economic value of tamarind (Tamarindus indica) in ecological zone I of Togo. Moroccan Journal of Agricultural and Veterinary Sciences. 10 : 272-281.
Samarou M., Atakpama W., Batawila K., Akpagana K. (2022b). State of knowledge on Tamarindus indica L. (Fabaceae). Agronomie Africaine SP. 34: 313–323.
Shekhar M., Singh N., Dutta R., Kumar S., Mahajan V. (2017). Comparative study of qualitative and quantitative methods to determine toxicity level of Aspergillus flavus isolates in maize. PLOS ONE. 12: e0189760. [DOI: 10.1371/journal.pone. 0189760]
Soloviev P., Niang T.D., Gaye A., Totte A. (2004). Variability of fruit physicochemical characters for three harvested woody species in Senegal: Adansonia digitata, Balanites aegyptiaca and Tamarindus indica. Fruits. 59: 109-119. [DOI : 10.1051/fruits:2004011]
Stege C.V.D., Prehsler S., Hartl A., Vogl C.R. (2011). Tamarind (Tamarindus indica L.) in the traditional West African diet: not just a famine food. Fruits. 66: 171–185. [DOI: 10.1051/ fruits/2011025]
Yao K.M., Kambire O., Coulibaly K.R., Diomande A., Koffi-nevry R. (2021). Microbiological and physicochemical quality of «Tomi»: an artisanal drink made from Tamarindus indica pulp, sold in Korhogo (Côte d’Ivoire). International Journal of Innovation and Applied Studies. 34: 551-560.
* Corresponding author (S. Aka-Gbezo)
* E-mail: solange.aka@csrs.ci
ORCID ID: https://orcid.org/0000-0002-9608-9139
* E-mail: solange.aka@csrs.ci
ORCID ID: https://orcid.org/0000-0002-9608-9139
Type of Study: Original article |
Subject:
Special
Received: 25/01/07 | Accepted: 25/07/22 | Published: 25/09/30
Received: 25/01/07 | Accepted: 25/07/22 | Published: 25/09/30
References
1. Abd El-Aziz A.R.M., Shehata S.M., Hisham S.M., Alobathani A.A. (2021). Molecular profile of aflatoxigenic and non-aflatoxigenic isolates of Aspergillus flavus isolated from stored maize. Saudi Journal of Biological Sciences. 28: 1383-1391. [DOI: 10.1016/j.sjbs.2020.11.073] [DOI:10.1016/j.sjbs.2020.11.073] [PMID] [PMCID]
2. Abdel-Hadi A., Schmidt-Heydt M., Parra R., Geisen R., Magan N. (2011). A systems approach to model the relationship between aflatoxin gene cluster expression, environmental factors, growth and toxin production by Aspergillus flavus. Journal of The Royal Society Interface. 9: 757-767. [DOI: 10.1098/rsif.2011.0482] [DOI:10.1098/rsif.2011.0482] [PMID] [PMCID]
3. Aka S., Yao K.F., Amoikon L.-S.T., Balla B., Bouatenin J.-P. (2025). Socio-economic, physico-chemical and microbiological study of tamarind pulps (Tamarindus indica) sold in Adjamé and Boundiali (Côte d'Ivoire). Moroccan Journal of Agricultural and Veterinary Sciences. 13: 19-26. [DOI: 10.5281/zenodo. 15005651]
4. Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. (1990). Basic local alignment search tool. Journal of Molecular Biology. 215: 403-410. [DOI: 10.1016/S0022-2836(05)80360-2]. available at http://blast.ncbi.nlm.nih.gov/Blast.cgi. [DOI:10.1016/S0022-2836(05)80360-2] [PMID]
5. Asoiro F.U., Ezeoha S.L., Ezenne G.I., Ugwu C.B. (2017). Chemical and mechanical properties of velvet tamarind fruit (Dialium guineense). Nigerian Journal of Technology. 36: 252-260. [DOI: 10.4314/njt.v36i1.30] [DOI:10.4314/njt.v36i1.30]
6. Bakayoko N.L., Soumahoro S., Tiekoura K.B., Ouattara M.B., Guede K.B., Coulibaly A.M., Guessennd N.K., Camara-Koussemon C (2024a). Diversity of lactic acid bacteria involved in the fermentation of the tamarind fruit (Tamarindus indica) Côte d'Ivoire. International Journal of Advanced Research. 12: 766-774. [DOI:10.21474/IJAR01/18938] [DOI:10.21474/IJAR01/18938]
7. Bakayoko N.L., Tiekoura K.B., Soumahoro S., Toure A., Guessennd N.K., Camara-Koussemon C (2024b). Food and therapeutic uses of tamarind fruit in Korhogo and Abidjan, Côte d'Ivoire. Afrique Science. 24: 47-59. [DOI:10.9734/jamb/2024/v24i12870]
8. Bian C., Kusuya Y., Sklenář F., D'hooge E., Yaguchi T., Ban S., Visagie C.M., Houbraken J., Takahashi H., Hubka V. (2022). Reducing the number of accepted species in Aspergillus series Nigri. Studies in Mycology. 102: 95-132. [DOI: 10.3114/sim.2022.102.03] [DOI:10.3114/sim.2022.102.03] [PMID] [PMCID]
9. Chanprasartsuk O., Prakitchaiwattana C., Sanguandeekul R. (2013). Comparison of methods for identification of yeasts isolated during spontaneous fermentation of freshly crushed pineapple juices. Journal of Agricultural Science and Technology. 15: 1779-1790. URL: http://jast.modares.ac.ir/article-23-5776-en.html.
10. Devi B., Boruah T. (2020). Tamarind (Tamarindus indica). In: Nayik G.A., Gull A. (Editors). Antioxidants in fruits: properties and health benefits. Springer Nature, Singapore. pp: 317-332. [DOI: 10.1007/978-981-15-7285-2_16] [DOI:10.1007/978-981-15-7285-2_16]
11. Diallo B.O., Joly H.I., Key D.M., Hossaert-Mckey M., Chevallier M.H. (2010). Change in biometric characters of seeds and seedlings of nine provenances of Tamarindus indica L. (Caesalpinioideae). Fruits. 65: 153-167. [DOI: 10.1051/fruits/2010010]. [DOI:10.1051/fruits/2010010]
12. Diguță C.F., Proca I.G., Jurcoane S. Matei F. (2019). Molecular characterization by PCR-RFLP of indigenous fungal isolates from hypersaline stream water in România. Folia Microbiological. 64: 407-414. [DOI: 10.1007/s12223-018-0664-6] [DOI:10.1007/s12223-018-0664-6] [PMID]
13. Diguta C.F., Vincent B., Guilloux-Benatier M., Alexandre H., Rousseaux S. (2011). PCR ITS-RFLP: a useful method for identifying filamentous fungi isolates on grapes. Food Microbiology. 28: 1145-1154. [DOI:10.1016/j.fm.2011.03.006] [DOI:10.1016/j.fm.2011.03.006] [PMID]
14. Erkmen O., Bozoglu T. F. (2016). Food preservation by reducing water activity. In: Erkmen O., Bozoglu T.F. (Editors). Food microbiology: principles into practice. John Wiley and Sons, Chichester, West Sussex. pp: 44-58. [DOI: 10.1002/9781119237860.ch30] [DOI:10.1002/9781119237860.ch30]
15. Frisvad J.C., Hubka V., Ezekiel C.N., Hong S.-B., Nováková A., Chen A.J., Arzanlou M., Larsen T.O., Sklenář F., Mahakarnchanakul W., Samson R.A., Houbraken J. (2019). Taxonomy of Aspergillus section Flavi and their production of aflatoxins, ochratoxins and other mycotoxins. Studies in Mycology. 93: 1-63. [DOI: 10.1016/j.simyco.2018.06.001] [DOI:10.1016/j.simyco.2018.06.001] [PMID] [PMCID]
16. Gacem M. (2011). Contribution to study of the antifungal and antimycotoxigenic activity of methanol and aqueous extract of seeds of Citrullus colocynthis against some fungi isolated from wheat stored. Doctoral dissertation. Kasdi Merbah University of Ouargla, Algeria. 149. URL: https://dspace.univ-ouargla.dz/ jspui/bitstream/123456789/487/1/GACEM_Mohamed_Amine.pdf.
17. Ghiasian S.A., Kord-Bacheh P., Rezayat S.M., Maghsood A.H., Taherkhani H. (2004). Mycoflora of Iranian maize harvested in the main production areas in 2000. Mycopathologia. 158: 113-121. [DOI: 10.1023/B:MYCO.0000038425.95049.03] [DOI:10.1023/B:MYCO.0000038425.95049.03] [PMID]
18. Grollier C., Debien C., Dornier M., Reynes M. (1998). Prominent characteristics and possible uses of the tamarind. Fruits. 53: 271-280.
19. Hamacek F.R., Santos P.R.G., Cardoso L.D.M., Pinheiro-Sant'Ana H.M. (2013). Nutritional composition of tamarind (Tamarindus indica L.) from the Cerrado of Minas Gerais, Brazil. Fruits. 68: 381-395. [DOI: 10.1051/fruits/2013083] [DOI:10.1051/fruits/2013083]
20. International Organization for Standardization (ISO). (1998). Fruit and vegetable products - determination of titratable acidity (ISO 750:1998). URL: https://cdn.standards.iteh.ai/samples/22569/eef01c3051f247398eb411358ccf25d1/ISO-750-1998.pdf.
21. Kabak B. (2009). The fate of mycotoxins during thermal food processing. Journal of the Science of Food and Agriculture. 89: 549-554. [DOI: 10.1002/jsfa.3491] [DOI:10.1002/jsfa.3491]
22. Kizis D., Natskoulis P., Nychas G.-J.E., Panagou E.Z. (2014). Biodiversity and ITS-RFLP characterisation of Aspergillus section Nigri isolates in grapes from four traditional grape-producing areas in Greece. PLOS ONE. 9: e93923. [DOI: 10.1371/journal.pone.0093923] [DOI:10.1371/journal.pone.0093923] [PMID] [PMCID]
23. Konan G.A.J., Nimaga D., Kouakou K.N., N'dri Y.D., Amani N.G. (2022). Identification of the main dishes made from the pulp of tomi (Tamarindus indica L) consumed in the Savannah Region of Côte d'Ivoire. Journal of Experimental Agriculture International. 44: 224-237. [DOI: 10.9734/JEAI/2022/v44i1030899] [DOI:10.9734/jeai/2022/v44i1030899]
24. Kouadio J.H., Lattanzio V.M.T., Ouattara D., Kouakou B., Visconti A. (2014). Assessment of mycotoxin exposure in Côte d'Ivoire (Ivory Coast) through multi-biomarker analysis and possible correlation with food consumption patterns. Toxicology International. 21: 248-257. [DOI: 10.4103/0971-6580.155336] [DOI:10.4103/0971-6580.155336] [PMID] [PMCID]
25. Lin J.-Q., Zhao X.-X., Wang C.-C., Xie Y., Li G.-H., He Z.-M. (2013). 5-Azacytidine inhibits aflatoxin biosynthesis in Aspergillus flavus. Annals of Microbiology. 63: 763-769. [DOI: 10.1007/s13213-012-0531-7] [DOI:10.1007/s13213-012-0531-7]
26. Mafe A.N., Büsselberg D. (2024). Mycotoxins in food: cancer risks and strategies for control. Foods. 13: 3502. [DOI: 10.3390/foods13213502] [DOI:10.3390/foods13213502] [PMID] [PMCID]
27. Musangi C.R., Juma B.S., Mukhebi D.W., Isoe E.M., Kibiti C.M., Mbinda W.M. (2024). Aspergillus population diversity and its role in aflatoxin contamination of cashew nuts from coastal Kenya. PLOS ONE. 19: e0292519. [DOI: 10.1371/journal.pone.0292519] [DOI:10.1371/journal.pone.0292519] [PMID] [PMCID]
28. Navale V., Vamkudoth K.R., Ajmera S., Dhuri V. (2021). Aspergillus derived mycotoxins in food and the environment: prevalence, detection, and toxicity. Toxicology Reports. 8: 1008-1030. [DOI: 10.1016/j.toxrep.2021.04.013] [DOI:10.1016/j.toxrep.2021.04.013] [PMID] [PMCID]
29. Nji N.Q., Christianah A.M., Njie A.C., Mulunda M. (2022). Biodiversity and distribution of Aspergillus and their toxins in maize from western and eastern regions of South Africa. Advances in Microbiology. 12: 121-149. [DOI : 10.4236/aim.2022.123011] [DOI:10.4236/aim.2022.123011]
30. Obulesu M., Bhattacharya S. (2011). Color changes of tamarind (Tamarindus indica L.) pulp during fruit development, ripening, and storage. International Journal of Food Properties. 14: 538-549. [DOI: 10.1080/10942910903262129]. [DOI:10.1080/10942910903262129]
31. Ouattara N.D., Gaille E., Stauffer F.W., Bakayoko A. (2016). Floristic diversity and ethnobotany of edible wild plants in the Department of Bondoukou (North-Eastern Côte d'Ivoire). Journal of Applied Biosciences. 98: 9284-9300. [DOI: 10.4314/jab.v98i1.5] [DOI:10.4314/jab.v98i1.5]
32. Rao K.R., Vipin A.V., Venkateswaran G. (2020). Molecular profile of non-aflatoxigenic phenotype in native strains of Aspergillus flavus. Archives of Microbiology. 202: 1143-1155. [DOI: 10.1007/s00203-020-01822-1] [DOI:10.1007/s00203-020-01822-1] [PMID]
33. Ronoh P.K., Toroitich F.J., Makonde H.M., Lelmen E.K., Obonyo M.A. (2024). Reliability of the chemical, metabolic, and molecular methods in discriminating aflatoxigenic from non-aflatoxigenic Aspergillus isolates. The Microbe. 4: 100115. [DOI: 10.1016/j.microb.2024.100115] [DOI:10.1016/j.microb.2024.100115]
34. Samarou M., Atakpama W., Atato A., Pessinaba Mamoudou M., Batawila K., Akpagana K. (2022a). Socio-economic value of tamarind (Tamarindus indica) in ecological zone I of Togo. Moroccan Journal of Agricultural and Veterinary Sciences. 10 : 272-281.
35. Samarou M., Atakpama W., Batawila K., Akpagana K. (2022b). State of knowledge on Tamarindus indica L. (Fabaceae). Agronomie Africaine SP. 34: 313-323.
36. Shekhar M., Singh N., Dutta R., Kumar S., Mahajan V. (2017). Comparative study of qualitative and quantitative methods to determine toxicity level of Aspergillus flavus isolates in maize. PLOS ONE. 12: e0189760. [DOI: 10.1371/journal.pone. 0189760] [DOI:10.1371/journal.pone.0189760] [PMID] [PMCID]
37. Soloviev P., Niang T.D., Gaye A., Totte A. (2004). Variability of fruit physicochemical characters for three harvested woody species in Senegal: Adansonia digitata, Balanites aegyptiaca and Tamarindus indica. Fruits. 59: 109-119. [DOI : 10.1051/fruits:2004011] [DOI:10.1051/fruits:2004011]
38. Stege C.V.D., Prehsler S., Hartl A., Vogl C.R. (2011). Tamarind (Tamarindus indica L.) in the traditional West African diet: not just a famine food. Fruits. 66: 171-185. [DOI: 10.1051/ fruits/2011025] [DOI:10.1051/fruits/2011025]
39. Yao K.M., Kambire O., Coulibaly K.R., Diomande A., Koffi-nevry R. (2021). Microbiological and physicochemical quality of «Tomi»: an artisanal drink made from Tamarindus indica pulp, sold in Korhogo (Côte d'Ivoire). International Journal of Innovation and Applied Studies. 34: 551-560. [DOI:10.1155/2021/5572300] [PMID] [PMCID]
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |