Volume 11, Issue 4 (December 2024)
J. Food Qual. Hazards Control 2024, 11(4): 280-290 |
Back to browse issues page
Ethics code: Not applicable
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Mall D, Patel V, Subhash R. Conventional and Molecular Characterization Based Microbial Assessment of Street Vended (Vada pav) Samples from Anand City, Gujarat, India. J. Food Qual. Hazards Control 2024; 11 (4) :280-290
URL: http://jfqhc.ssu.ac.ir/article-1-1210-en.html
URL: http://jfqhc.ssu.ac.ir/article-1-1210-en.html
Post Graduate Department of Home Science, Sardar Patel University, Vallabh Vidyanagar-388120, Gujarat, India , patelvh2004@yahoo.co.in
Full-Text [PDF 561 kb]
(373 Downloads)
| Abstract (HTML) (1241 Views)
Full-Text: (471 Views)
Conventional and Molecular Characterization Based Microbial Assessment of Street Vended (Vada pav) Samples from Anand City, Gujarat, India
D.P. Mall, V.H. Patel [*]* , R. Subhash
Post Graduate Department of Home Science, Sardar Patel University, Vallabh Vidyanagar-388120, Gujarat, India
HIGHLIGHTS
Table 1: Details of primers used for Polymerase Chain Reaction (PCR) assay
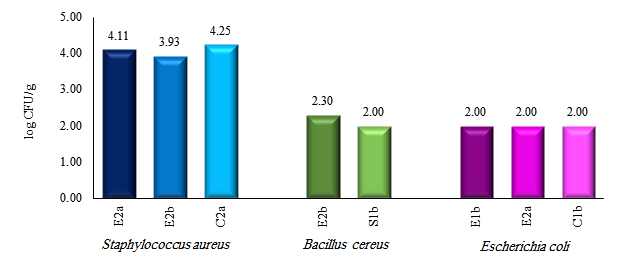
Figure 1: Bacterial counts of different Vada pav samples
CFU=Colony Forming Units, first two letters=location, a and b=trial
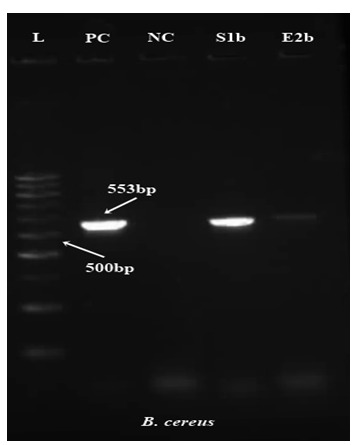
Figure 2: Polymerase Chain Reaction (PCR) amplification of nheA gene for Bacillus cereus and phoA gene for Escherichia coli isolates obtained from Vada pav samples from different locations
L=DNA Ladder (100bp), PC=Positive Control, NC=Negative Control, E 1 and E2=East 1 and East 2, S1=South 1, C1=Central 1, a and b=trial
Table 4: Microbial contamination of Vada pav samples by location
CFU=Colony Forming Units; TVC=Total viable count; YMC=Yeast and Mold Count; P=Positive; *N=Negative result by PCR assay; E1 and E2=East zone; W1=West zone; N1=North zone; S1=South zone; C1 and C2=Central zone; a and b=trial
Table 5: Comparison of microbial conventional and molecular methods in Vada pav samples (n=7)
D.P. Mall, V.H. Patel [*]*
Post Graduate Department of Home Science, Sardar Patel University, Vallabh Vidyanagar-388120, Gujarat, India
- The total viable count in Vada pav samples ranged from 3.94 to 6.09 CFU/g, while the Yeast and Mold Count ranged from 1.15 to 2.85 log CFU/g tested in seven different locations.
- The dominant microbes identified in Vada pav sample, using both methods, were Escherichia coli (42.85%) followed by Bacillus cereus (28.5%).
- Staphylococcus aureus (28.5%) was isolated by the conventional culture technique but not confirmed using molecular characterization.
- Salmonella spp. was not detected in any of the samples.
| Article type Original article |
ABSTRACT Background: Street foods offer convenient meal options for the consumer, but pose safety concerns if not handled or served with proper hygiene. The purpose of the present study was the microbial evaluation of street vended Vada pav samples sold at popular locations in Anand city using the conventional culture technique and molecular characterization via Polymerase Chain Reaction. Methods: Duplicate samples were collected from seven different locations (n=14) across five zones: East (2), West (1), North (1), South (1), and Central (2) during June 2023. Staphylococcus aureus, Bacillus cereus, Escherichia coli, and Salmonella spp. were isolated and identified. For the microbial screening, bacterial enumeration, colony morphology, Gram's reaction, and biochemical characterization were performed. Amplification of nuc (S. aureus), nheA (B. cereus), phoA (E. coli), and 16S rRNA (Salmonella spp.) genes were carried out via Polymerase Chain Reaction assay. Results: Total Viable Count (TVC) ranged from 3.93 to 6.08 log Colony Forming Units (CFU)/g while the Yeast and Mold Counts ranged from 2.30 to 4.28 log CFU/g. Using the conventional culture technique, the prevalence of S. aureus, B. cereus, and E. coli were found to be 3/14, 2/14, and 3/14, respectively; whereas based on molecular characterization, the prevalence was 0/14, 2/14, and 3/14, respectively. Salmonella spp. was not detected in any of the samples. Conclusion: The study indicates a potential health hazard for consumers due to microbial contamination of street vended Vada pav samples. Consequently, it is crucial to regulate and improve hygienic practices in street food vendors. © 2024, Shahid Sadoughi University of Medical Sciences. This is an open access article under the Creative Commons Attribution 4.0 International License. |
|
| Keywords Molecular Typing Staphylococcus aureus Polymerase Chain Reaction Food Safety India |
||
| Article history Received: 31 Mar 2024 Revised: 03 Jul 2024 Accept: 10 Nov 2024 |
||
| Abbreviations CFU=Colony Forming Units IMViC=Indole, Methyl Red, Voges–Proskauer, and Citrate MR=Methyl Red PCR=Polymerase Chain Reaction TVC=Total Viable Count VP=Voges Proskauer YMC=Yeast and Mold Count |
To cite: Mall D.P., Patel V.H., Subhash R. (2024). Conventional and molecular characterization based microbial assessment of street vended (Vada pav ) samples from Anand city, Gujarat, India. Journal of Food Quality and Hazards Control. 11: 280-290.
Introduction
Introduction
Street Vended Foods (SVFs) are readily available meals for the consumer. India is famous for its unique street foods which contribute up to 40% of the daily diet of the urban population (Hirani et al., 2019). Street foods are popular among city dwellers because they are inexpensive, easily accessible, and convenient on the sidewalk, especially for low to middle income populations (Alelign et al., 2023). Vada pav is a popular Indian street food that consists of a spicy mashed potato patty called Vada, which is deep-fried after being coated in chickpea batter. This vada is placed inside a soft bread roll called pav, usually topped with various chutneys or sauces for added flavour (Solomon et al., 2015). The safety of these foods is always doubtful. These foods are perceived as potential risks for food-borne illnesses if not prepared or served hygienically. According to WHO (2022) estimates, unsafe food is responsible for 420,000 deaths and 600 million incidents of food-borne illnesses globally each year. Children under the age of five account for 30% of food-borne deaths. Eating unsafe food results in the loss of 33 million years of healthy life annually worldwide; however, this figure is probably underestimated. Staphylococcus aureus, Bacillus cereus, Salmonella, Campylobacter jejuni, Listeria monocytogenes, Escherichia coli, Clostridium botulinum, C. perfringens, Shigella, and Vibrio are food-borne pathogens that cause several food-borne illnesses (FSSAI, 2021).
The second or third most common cause of food-borne illnesses is S. aureus. Food products are considered one of the important sources from which this bacterium can infect humans and cause illness in both animals and humans. S. aureus demonstrates various pathogenic factors that contribute to its pathogenicity and ability to colonize (Rajabi et al., 2023). Food contamination with enterotoxigenic S. aureus leads to staphylococcal enterotoxin intoxication, resulting in acute symptoms such as nausea, vomiting, abdominal cramps, diarrhoea, and chills, with or without fever (Fisher et al., 2018). B. cereus is an opportunistic pathogen, ranked as the fifth most common bacterium responsible for foodborne infections and food poisoning. It is widely distributed in the environment and is undoubtedly the most significant among aerobic spore forming species. B. cereus has the capacity to produce an emetic toxin and diarrheal enterotoxins (Hefny et al., 2020). E. coli typically inhabits the intestinal tract of vertebrate animals. Therefore, the identification of E. coli in food suggests the potential for faecal contamination and the presence of additional faecal bacteria, including pathogens. E. coli is one of the most effective faecal contamination indicator bacteria among commonly used faecal indicator bacteria (Braz et al., 2020). Salmonella spp. are found in both domestic and wild animals as well as in the environment. Salmonella spp. cause salmonellosis, a common food-borne infection in humans. Salmonella exposure can cause mild symptoms, severe disease, and even death (Food Standards Australia New Zealand, 2023).
Traditional microbiological methods, such as International Organisation for Standardisation (ISO) standards provide reliable and standardised procedures for detecting food-borne pathogens. However, enrichment, isolation, and identification steps need to be followed, which can lead to time consuming analysis that may not be compatible with the requirement for rapid results (Foddai and Grant., 2020). Rapid methods for identifying food-borne pathogens, including immunological, biosensor, and nucleic acid-based techniques, have been developed to address the limitations of traditional methods (Kim et al., 2020). Polymerase Chain Reaction (PCR) is the most widely used nucleic acid amplification technique to detect food-borne pathogens. PCR assay detects pathogens by amplifying target genes using primers which correspond to a certain base sequences present in a microorganism however, electrophoresis is required to confirm both the presence and size of the desired final amplified product (Zhao et al., 2014).
The nuc gene encodes the thermonuclease enzyme and can be used as a valuable genetic marker for the identification of S. aureus using PCR (Al-Ashmawy et al., 2016). B. cereus produces enterotoxins such as cytotoxin K (CytK), non hemolytic enterotoxin (Nhe), and hemolysis BL (Hbl), which cause food-borne poisoning. The NHE gene complex consists of NheA, NheB, and NheC. The non-haemolytic enterotoxin (Nhe) is responsible for the diarrhoeal food-poisoning syndrome (Liu et al., 2020). The alkaline phosphatase encoding gene phoA is a housekeeping gene present in all E. coli and can be utilised for specific identification (Luo et al., 2023). Salmonella spp. are amplified for the 16S rRNA gene because it is the most prevalent housekeeping genetic marker due to its presence across bacterial species (Ibal et al., 2019).
A few studies have been conducted in Gujarat state on the microbiological hazards of street vended foods. These studies were focused on pizza (Solanki and Dave, 2012) Pani puri (Mehta et al., 2020), Bhel (Sheth et al., 2005), and hotdogs (Jotangiya, 2018). Further, based on the literature reviewed, no studies have been reported from Anand city on street vended foods. Vada pav is a popular ready-to-eat food; however, its microbial contamination has not been reported from any part of India except for a study by Chumber et al. (2007) from Pune. Thus, the purpose of this study was to carry out a microbial evaluation of street vended Vada pav samples sold in popular locations in Anand city of central Gujarat with reference to S. aureus, B. cereus, E. coli, and Salmonella spp.
Materials and methods
Sample collection
Samples of Vada pav were purposively collected in June 2023 in two trials (a and b) from seven popular locations out of a total of nine in Anand city, as follows: two from the east (E1 and E2), one from the west (W1), one from the north (N1), one from the south (S1), and two from the central zone (C1 and C2). Samples were brought to the laboratory in plastic bags as sold by the vendor, along with tomato sauce, with or without added chutney in a separate smaller plastic bag and then transported in an ice box and processed within 2 h.
Sample preparation
In the laboratory, under aseptic conditions, Vada pav was carefully opened, the pav (bread) was separated and a small quantity (2 g) of tomato sauce or a mixture of tomato sauce and chutney was spread aseptically on the Vada, as Vada pav is generally consumed with tomato sauce and chutney. The Vada pav was reassembled according to the vendor's presentation. For sampling, four small pieces of Vada pav were collected from four different sides, along with a portion from the centre. All pieces were combined and weighed to obtain a total weight of 25 g. This sample was placed inside a sterile filtered sampling bag (PW391, HiMedia) with the addition of 225 ml of Buffered Peptone Water (BPW; M1275, HiMedia). A homogenized mixture was prepared by crushing it with a pestle (for up to one min). Since the sampling bag contains a filter, the peptone water passes through to the opposite side while the homogenized solid material remains inside. The BPW obtained was further serially diluted up to 10-3 decimal dilutions. For plating, 100 µl of the sample was pipetted out and spread onto basal as well as selective and differential media.
Conventional culture technique
-Identification and enumeration of bacteria
Samples were inoculated on total plate count agar (M091A, HiMedia, Mumbai, India) and potato dextrose agar (M096, HiMedia,Mumbai, India) for Total Viable Count (TVC) and Yeast and Mold Count (YMC), respectively. Baird Parker Agar (BPA; M043,HiMedia,Mumbai, India) containing 5% egg yolk tellurite emulsion (FD046), Polymyxin Pyruvate Egg Yolk Mannitol Bromothymol Blue Agar (PEMBA; M1484, HiMedia,Mumbai, India) with the addition of egg yolk emulsion (5%) (FD045, HiMedia,Mumbai, India) and PEMBA supplement (5%) (FD200, HiMedia,Mumbai, India), Hicrome E. coli Agar (HEA; M1295I HiMedia,Mumbai, India), and Salmonella Shigella Agar modified (SSA; M1032, HiMedia,Mumbai, India) were used for the identification of S. aureus, B. cereus, E. coli, and Salmonella species, respectively. Plates were incubated at 37 oC for 24 to 48 h and colonies were counted and replated on respective media for isolation. Results were expressed in log Colony Forming Units (CFU)/g. Isolated bacteria were characterized based on colony morphology, Gram’s reaction, motility test, and biochemical tests. For biochemical tests, carbohydrate utilization (glucose, lactose, sucrose, mannose, arabinose, dulcitol, and mannitol), Indole, Methyl Red, Voges–Proskauer, and Citrate (IMViC), hemolysin production; Triple Sugar Iron agar (TSI) test; and enzyme production test such as catalase, oxidase, coagulase, nitratase, urease; as well as the hydrolysis of starch, gelatin, and casein were performed (Patel and Patel, 2016). Media and reagents for phenotypic characterization were purchased from HiMedia (Mumbai, India) and prepared in the laboratory.
-Reference strains
All bacterial strains used as positive controls, S. aureus - 7443, B. cereus (6840), E. coli – (1692), and Salmonella spp. (734) were purchased from the Microbial Type Culture Collection and Gene Bank (MTCC) (Chandigarh, India).
Molecular characterisation using PCR assay
-DNA extraction
DNA was extracted using the thermal lysis method. Pure cultures were suspended in 100 μl of nuclease free water in a sterile microcentrifuge tube. The bacterial suspension was vortexed and heated in a thermal cycler (Applied biosystems, California- 2720) at 95 °C for 10 min. The samples were centrifuged at 10,000 rpm for 10 min to settle the cell debris. The 50 ul upper aqueous phase containing bacterial DNA was transferred to another microcentrifuge tube and stored at -20 oC until further use (Likhitha et al., 2022).
-PCR protocol
Presumptive isolates were subjected to PCR assay for species specific thermonuclease gene nuc for S. aureus, the diarrheal enterotoxin gene nheA for B. cereus, the housekeeping gene phoA for E. coli, and 16S rRNA for Salmonella spp., respectively. Table 1 provides details on the oligonucleotide primers used for all four genes. The reaction mixture was prepared according to the method outlined by Thakur et al. (2020)
The second or third most common cause of food-borne illnesses is S. aureus. Food products are considered one of the important sources from which this bacterium can infect humans and cause illness in both animals and humans. S. aureus demonstrates various pathogenic factors that contribute to its pathogenicity and ability to colonize (Rajabi et al., 2023). Food contamination with enterotoxigenic S. aureus leads to staphylococcal enterotoxin intoxication, resulting in acute symptoms such as nausea, vomiting, abdominal cramps, diarrhoea, and chills, with or without fever (Fisher et al., 2018). B. cereus is an opportunistic pathogen, ranked as the fifth most common bacterium responsible for foodborne infections and food poisoning. It is widely distributed in the environment and is undoubtedly the most significant among aerobic spore forming species. B. cereus has the capacity to produce an emetic toxin and diarrheal enterotoxins (Hefny et al., 2020). E. coli typically inhabits the intestinal tract of vertebrate animals. Therefore, the identification of E. coli in food suggests the potential for faecal contamination and the presence of additional faecal bacteria, including pathogens. E. coli is one of the most effective faecal contamination indicator bacteria among commonly used faecal indicator bacteria (Braz et al., 2020). Salmonella spp. are found in both domestic and wild animals as well as in the environment. Salmonella spp. cause salmonellosis, a common food-borne infection in humans. Salmonella exposure can cause mild symptoms, severe disease, and even death (Food Standards Australia New Zealand, 2023).
Traditional microbiological methods, such as International Organisation for Standardisation (ISO) standards provide reliable and standardised procedures for detecting food-borne pathogens. However, enrichment, isolation, and identification steps need to be followed, which can lead to time consuming analysis that may not be compatible with the requirement for rapid results (Foddai and Grant., 2020). Rapid methods for identifying food-borne pathogens, including immunological, biosensor, and nucleic acid-based techniques, have been developed to address the limitations of traditional methods (Kim et al., 2020). Polymerase Chain Reaction (PCR) is the most widely used nucleic acid amplification technique to detect food-borne pathogens. PCR assay detects pathogens by amplifying target genes using primers which correspond to a certain base sequences present in a microorganism however, electrophoresis is required to confirm both the presence and size of the desired final amplified product (Zhao et al., 2014).
The nuc gene encodes the thermonuclease enzyme and can be used as a valuable genetic marker for the identification of S. aureus using PCR (Al-Ashmawy et al., 2016). B. cereus produces enterotoxins such as cytotoxin K (CytK), non hemolytic enterotoxin (Nhe), and hemolysis BL (Hbl), which cause food-borne poisoning. The NHE gene complex consists of NheA, NheB, and NheC. The non-haemolytic enterotoxin (Nhe) is responsible for the diarrhoeal food-poisoning syndrome (Liu et al., 2020). The alkaline phosphatase encoding gene phoA is a housekeeping gene present in all E. coli and can be utilised for specific identification (Luo et al., 2023). Salmonella spp. are amplified for the 16S rRNA gene because it is the most prevalent housekeeping genetic marker due to its presence across bacterial species (Ibal et al., 2019).
A few studies have been conducted in Gujarat state on the microbiological hazards of street vended foods. These studies were focused on pizza (Solanki and Dave, 2012) Pani puri (Mehta et al., 2020), Bhel (Sheth et al., 2005), and hotdogs (Jotangiya, 2018). Further, based on the literature reviewed, no studies have been reported from Anand city on street vended foods. Vada pav is a popular ready-to-eat food; however, its microbial contamination has not been reported from any part of India except for a study by Chumber et al. (2007) from Pune. Thus, the purpose of this study was to carry out a microbial evaluation of street vended Vada pav samples sold in popular locations in Anand city of central Gujarat with reference to S. aureus, B. cereus, E. coli, and Salmonella spp.
Materials and methods
Sample collection
Samples of Vada pav were purposively collected in June 2023 in two trials (a and b) from seven popular locations out of a total of nine in Anand city, as follows: two from the east (E1 and E2), one from the west (W1), one from the north (N1), one from the south (S1), and two from the central zone (C1 and C2). Samples were brought to the laboratory in plastic bags as sold by the vendor, along with tomato sauce, with or without added chutney in a separate smaller plastic bag and then transported in an ice box and processed within 2 h.
Sample preparation
In the laboratory, under aseptic conditions, Vada pav was carefully opened, the pav (bread) was separated and a small quantity (2 g) of tomato sauce or a mixture of tomato sauce and chutney was spread aseptically on the Vada, as Vada pav is generally consumed with tomato sauce and chutney. The Vada pav was reassembled according to the vendor's presentation. For sampling, four small pieces of Vada pav were collected from four different sides, along with a portion from the centre. All pieces were combined and weighed to obtain a total weight of 25 g. This sample was placed inside a sterile filtered sampling bag (PW391, HiMedia) with the addition of 225 ml of Buffered Peptone Water (BPW; M1275, HiMedia). A homogenized mixture was prepared by crushing it with a pestle (for up to one min). Since the sampling bag contains a filter, the peptone water passes through to the opposite side while the homogenized solid material remains inside. The BPW obtained was further serially diluted up to 10-3 decimal dilutions. For plating, 100 µl of the sample was pipetted out and spread onto basal as well as selective and differential media.
Conventional culture technique
-Identification and enumeration of bacteria
Samples were inoculated on total plate count agar (M091A, HiMedia, Mumbai, India) and potato dextrose agar (M096, HiMedia,Mumbai, India) for Total Viable Count (TVC) and Yeast and Mold Count (YMC), respectively. Baird Parker Agar (BPA; M043,HiMedia,Mumbai, India) containing 5% egg yolk tellurite emulsion (FD046), Polymyxin Pyruvate Egg Yolk Mannitol Bromothymol Blue Agar (PEMBA; M1484, HiMedia,Mumbai, India) with the addition of egg yolk emulsion (5%) (FD045, HiMedia,Mumbai, India) and PEMBA supplement (5%) (FD200, HiMedia,Mumbai, India), Hicrome E. coli Agar (HEA; M1295I HiMedia,Mumbai, India), and Salmonella Shigella Agar modified (SSA; M1032, HiMedia,Mumbai, India) were used for the identification of S. aureus, B. cereus, E. coli, and Salmonella species, respectively. Plates were incubated at 37 oC for 24 to 48 h and colonies were counted and replated on respective media for isolation. Results were expressed in log Colony Forming Units (CFU)/g. Isolated bacteria were characterized based on colony morphology, Gram’s reaction, motility test, and biochemical tests. For biochemical tests, carbohydrate utilization (glucose, lactose, sucrose, mannose, arabinose, dulcitol, and mannitol), Indole, Methyl Red, Voges–Proskauer, and Citrate (IMViC), hemolysin production; Triple Sugar Iron agar (TSI) test; and enzyme production test such as catalase, oxidase, coagulase, nitratase, urease; as well as the hydrolysis of starch, gelatin, and casein were performed (Patel and Patel, 2016). Media and reagents for phenotypic characterization were purchased from HiMedia (Mumbai, India) and prepared in the laboratory.
-Reference strains
All bacterial strains used as positive controls, S. aureus - 7443, B. cereus (6840), E. coli – (1692), and Salmonella spp. (734) were purchased from the Microbial Type Culture Collection and Gene Bank (MTCC) (Chandigarh, India).
Molecular characterisation using PCR assay
-DNA extraction
DNA was extracted using the thermal lysis method. Pure cultures were suspended in 100 μl of nuclease free water in a sterile microcentrifuge tube. The bacterial suspension was vortexed and heated in a thermal cycler (Applied biosystems, California- 2720) at 95 °C for 10 min. The samples were centrifuged at 10,000 rpm for 10 min to settle the cell debris. The 50 ul upper aqueous phase containing bacterial DNA was transferred to another microcentrifuge tube and stored at -20 oC until further use (Likhitha et al., 2022).
-PCR protocol
Presumptive isolates were subjected to PCR assay for species specific thermonuclease gene nuc for S. aureus, the diarrheal enterotoxin gene nheA for B. cereus, the housekeeping gene phoA for E. coli, and 16S rRNA for Salmonella spp., respectively. Table 1 provides details on the oligonucleotide primers used for all four genes. The reaction mixture was prepared according to the method outlined by Thakur et al. (2020)
Table 1: Details of primers used for Polymerase Chain Reaction (PCR) assay
| Bacteria | Target Gene | Primer Sequence (5′–3′) | Amplicon size (bp) | GenBank accession number | Reference |
| Staphylococcus aureus | nuc | F: GCGATTGATGGTGATACGGTT | 270 | NC_007795.1 | Budiarso et al. (2019) |
| R: GCCAAGCCTTGACGAACTAAAGC | |||||
| Bacillus cereus | nheA | F: AAGGCGAATGTACGAGAGTGG | 553 | NZ_CP017060.1 | Kumar et al. (2009) |
| R: CTTCTCTCGTTTGACTATCTGCAG | |||||
| Escherichia coli | phoA | F: CGATTCTGGAAATGGCAAAAG | 720 | NC_002695.2 | Hu et al. (2011) |
| R: CGTGATCAGCGGTGACTATGAC | |||||
| Salmonella spp. | 16S rRNA | F: TGTTGTGGTTAATAACCGCA | 572 | NC_003197.2 | Nyabundi et al. (2015) |
| R: CACAAATCCATCTCTGGA |
-PCR conditions for the detection of the various genes
For S. aureus, the nuc gene was targeted using an initial denaturation at 94 °C for 5 min, followed by 35 cycles of denaturation at 95 °C for one min, annealing at 58 °C for 30 s, and extension at 72 °C for 45 s. A final extension was carried out at 72 °C for seven min. For B. cereus, the nheA gene was amplified with an initial denaturation at 94 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for one min, annealing at 61°C for 30 s, and extension at 72 °C for one min. For E. coli, the phoA gene was targeted with an initial denaturation at 94 °C for 5 min, followed by 30 cycles of denaturation at 94 °C for 45 s, annealing at 59 °C for 45 s, and extension at 72 °C for 45 s. For Salmonella spp., the 16S rRNA gene was amplified with an initial denaturation at 94 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 49 °C foronemin, and extension at 72 °C foronemin. A final extension step was performed at 72 °C for 7 min. The amplified product was subjected to gel electrophoresis on a 2% agarose gel and visualized using the Syngene Gbox gel documentation system (United kingdom).
Statistical analysis
One-way ANOVA was performed using IBM SPSS version 26.0 to determine significant differences (p≤0.05), with results presented as mean±Standard Deviation (SD) for TVC and YMC.
Results and discussion
Conventional culture technique
-Enumeration of TVC and YMC
Table 2 shows the TVC and YMC of Vada pav samples. TVC ranged from 3.94 to 6.09 log CFU/g, with the highest count found in a sample procured from one of the eastern zones (E1) and the lowest observed in the western zone (W3). YMC ranged from 1.15 to 2.85 log CFU/g with the highest YMC observed in the central zone (C1) and the lowest in one of the eastern zones (E2). Three locations (E1, N1, and S1) showed no yeast and mold contamination in the Vada pav samples. L4 and L5 showed low TVC and no contamination detected for YMC. TVC was significantly different (p≤0.05); however, YMC did not show significant differences among the analyzed Vada pav samples.
Table 2: Results of microbial counts in Vada pav samples (log Colony Forming Units (CFU)/g)
For S. aureus, the nuc gene was targeted using an initial denaturation at 94 °C for 5 min, followed by 35 cycles of denaturation at 95 °C for one min, annealing at 58 °C for 30 s, and extension at 72 °C for 45 s. A final extension was carried out at 72 °C for seven min. For B. cereus, the nheA gene was amplified with an initial denaturation at 94 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for one min, annealing at 61°C for 30 s, and extension at 72 °C for one min. For E. coli, the phoA gene was targeted with an initial denaturation at 94 °C for 5 min, followed by 30 cycles of denaturation at 94 °C for 45 s, annealing at 59 °C for 45 s, and extension at 72 °C for 45 s. For Salmonella spp., the 16S rRNA gene was amplified with an initial denaturation at 94 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 49 °C foronemin, and extension at 72 °C foronemin. A final extension step was performed at 72 °C for 7 min. The amplified product was subjected to gel electrophoresis on a 2% agarose gel and visualized using the Syngene Gbox gel documentation system (United kingdom).
Statistical analysis
One-way ANOVA was performed using IBM SPSS version 26.0 to determine significant differences (p≤0.05), with results presented as mean±Standard Deviation (SD) for TVC and YMC.
Results and discussion
Conventional culture technique
-Enumeration of TVC and YMC
Table 2 shows the TVC and YMC of Vada pav samples. TVC ranged from 3.94 to 6.09 log CFU/g, with the highest count found in a sample procured from one of the eastern zones (E1) and the lowest observed in the western zone (W3). YMC ranged from 1.15 to 2.85 log CFU/g with the highest YMC observed in the central zone (C1) and the lowest in one of the eastern zones (E2). Three locations (E1, N1, and S1) showed no yeast and mold contamination in the Vada pav samples. L4 and L5 showed low TVC and no contamination detected for YMC. TVC was significantly different (p≤0.05); however, YMC did not show significant differences among the analyzed Vada pav samples.
Table 2: Results of microbial counts in Vada pav samples (log Colony Forming Units (CFU)/g)
| Location | *TVC | *YMC |
| E1 | 6.09±0.32 b | *ND |
| E2 | 5.11±0.04 b | 1.15±1.63 a |
| W1 | 3.94±0.64 a | 2.14±3.03 a |
| N1 | 4.12±0.37 a | ND |
| S1 | 4.36±0.32 ab | ND |
| C1 | 4.36±0.14 ab | 2.85±0.52 a |
| C2 | 4.17±0.42 a | 1.35±1.91 a |
| F-value | 8.410 * | 1.162 |
ND=Not Detected; TV=Total Viable Count; YMC=Yeast and Mold Count
*=Significant difference (p≤0.05); Values are mean±Standard Deviation (SD) of 2 trials; means carrying similar superscripts within a column are not significantly different.
E1 and E2=East zone; W1=West zone; N1=North zone; S1=South zone; C1 and C2=Central zone; a and b=trial
Sharma et al. (2020) studied the total viable count of selected street foods, i.e., tikki, veggie burger, samosa, pakoda, momo, and spring rolls in Palampur city, Himachal Pradesh, India. They reported a TVC of 32×102 CFU/g for burgers, which is similar to the TVC obtained for all locations in the present study except E1 and E2. Asiegbu et al. (2020) studied the microbial quality of ready-to-eat street vended foods namely starch-based foods (n=75), beef-based foods (n=45), poultry-based foods (n=30), fish-based foods (n=10), vegetable-based foods (n=20), and sandwich-based foods (n=25). A count of 3.95±0.45 log CFU/g TVC was noted for sandwich-based foods; within this category, the highest count were observed for cheese burgers (n=5) and cheese/egg burgers (n=5) which was 4.12±0.28 and 3.77±0.30 CFU/g, respectively, which is close to the value obtained in the present investigation.
-Enumeration of S. aureus, B. cereus, E. coli and Salmonella spp.
Colonies observed on BPA exhibited a grey-black shiny appearance with or without an opaque zone surrounding the colony, indicative of S. aureus. PEMBA revealed colonies displaying a blue colour with precipitation, characteristic of B. cereus. On HEA agar, colonies displayed a bluish-green hue, indicating the presence of E. coli. Colonies of Salmonella spp. were not observed in any of the samples. The count for S. aureus ranged from 3.93 to 4.25 log CFU/g, while B. cereus count ranged from 2.00 to 2.30 log CFU/g. The count for E. coli was consistent at 2.00 log CFU/g, with the highest count of S. aureus found in samples obtained from the central zone. Samples acquired from the East and South zones were contaminated with B. cereus, while E. coli was isolated from the samples collected from both eastern and central zones (Figure 1).
*=Significant difference (p≤0.05); Values are mean±Standard Deviation (SD) of 2 trials; means carrying similar superscripts within a column are not significantly different.
E1 and E2=East zone; W1=West zone; N1=North zone; S1=South zone; C1 and C2=Central zone; a and b=trial
Sharma et al. (2020) studied the total viable count of selected street foods, i.e., tikki, veggie burger, samosa, pakoda, momo, and spring rolls in Palampur city, Himachal Pradesh, India. They reported a TVC of 32×102 CFU/g for burgers, which is similar to the TVC obtained for all locations in the present study except E1 and E2. Asiegbu et al. (2020) studied the microbial quality of ready-to-eat street vended foods namely starch-based foods (n=75), beef-based foods (n=45), poultry-based foods (n=30), fish-based foods (n=10), vegetable-based foods (n=20), and sandwich-based foods (n=25). A count of 3.95±0.45 log CFU/g TVC was noted for sandwich-based foods; within this category, the highest count were observed for cheese burgers (n=5) and cheese/egg burgers (n=5) which was 4.12±0.28 and 3.77±0.30 CFU/g, respectively, which is close to the value obtained in the present investigation.
-Enumeration of S. aureus, B. cereus, E. coli and Salmonella spp.
Colonies observed on BPA exhibited a grey-black shiny appearance with or without an opaque zone surrounding the colony, indicative of S. aureus. PEMBA revealed colonies displaying a blue colour with precipitation, characteristic of B. cereus. On HEA agar, colonies displayed a bluish-green hue, indicating the presence of E. coli. Colonies of Salmonella spp. were not observed in any of the samples. The count for S. aureus ranged from 3.93 to 4.25 log CFU/g, while B. cereus count ranged from 2.00 to 2.30 log CFU/g. The count for E. coli was consistent at 2.00 log CFU/g, with the highest count of S. aureus found in samples obtained from the central zone. Samples acquired from the East and South zones were contaminated with B. cereus, while E. coli was isolated from the samples collected from both eastern and central zones (Figure 1).

Figure 1: Bacterial counts of different Vada pav samples
CFU=Colony Forming Units, first two letters=location, a and b=trial
Yogesh et al. (2019) examined 30 burger samples from moderate fast food centres as well as reputed brands and street vendors and found significant contamination by Staphylococcus spp., E. coli, Salmonella spp., Vibrio spp., and Listeria spp. Adhikari et al. (2023) analyzed 150 chutney samples served at street foods sold in Bharatpur city, Nepal, reportingaverage CFU counts of 1.33×106, 1.83×105, and 1.24×105 for TVC, coliform, and Salmonella-Shigella counts, respectively. This indicates that chutneys used in Vada pav samples could be a source of microbial contamination. In the present investigation, a higher E. coli prevalence (42.85%) was observed compared to previous studies. Bezerra et al. (2010) evaluated hamburgers from Brazil and found contamination by Staphylococcus, B. cereus, and Salmonella spp. These authors noted high counts of Staphylococcus and B. cereus, but Salmonella spp. was not detected. The present study aligns with findings by Bezerra et al. (2010) as they also reported an absence of Salmonella spp.
Based on the FSSAI (2023) guidelines for food grain products such as bread, cakes, and doughnuts, the presence of Salmonella spp. is prohibited in these foods, which align with the findings of the present study. The microbial guidelines for ready-to-eat foods peovided by the Centre for Food Safety (2014) indicate that counts of S. aureus and E. coli should be <20, while B. cereus should be <103/g. However, the UKHSA (2024) has established a higher threshold for the presence of B. Cereus, set at <105. In the present study, the counts for S. aureus and E. coli exceeded the limit, while the count for B. cereus was lower.
Isolating Salmonella spp. from food is sometimes susceptible to failuredue to the loss of bacteria during enrichment, despite obtaining a contaminated portion. Salmonella spp. have the capability to enter a Viable but Non-Culturable (VBNC) state under unfavourable conditions, which adds to the challenges of culturing using various conventional culture media and enrichment protocols proposed for isolating Salmonella spp. from food and environmental samples (Pui et al., 2011).
-Morphological and biochemical characteristics
Subcultured isolates were examined for Gram's reaction and motility. Isolates of S. aureus were identified as Gram-positive cocci occurring singly or in diplo or staphylo arrangements. B. cereus isolates were confirmed as positive, rod-shaped bacteria occurring in short and long chains. Meanwhile, E. coli isolates exhibited Gram-negative, rod-shaped characteristics occurring singly or in diplo as observed under the microscope. In the motility test, S. aureus was found to be negative. All strains of B. cereus and E. coli demonstrated a diffuse cloud of growth away from the line of inoculation, indicating that all the strains were motile.
-IMViC test
Table 3 demonstrates the biochemical characteristics of the isolated bacteria. IMViC test showed that all S. aureus and B. cereus isolates tested negative for indole production and citrate utilisation. S. aureus isolates were positive for Methyl Red (MR) while B. cereus showed variable results. Variable results were also observed among S. aureus and B. cereus for Voges Proskauer (VP). Indole production and MR were positive for E. coli isolates, while VP and citrate utilisation were negative.
Based on the FSSAI (2023) guidelines for food grain products such as bread, cakes, and doughnuts, the presence of Salmonella spp. is prohibited in these foods, which align with the findings of the present study. The microbial guidelines for ready-to-eat foods peovided by the Centre for Food Safety (2014) indicate that counts of S. aureus and E. coli should be <20, while B. cereus should be <103/g. However, the UKHSA (2024) has established a higher threshold for the presence of B. Cereus, set at <105. In the present study, the counts for S. aureus and E. coli exceeded the limit, while the count for B. cereus was lower.
Isolating Salmonella spp. from food is sometimes susceptible to failuredue to the loss of bacteria during enrichment, despite obtaining a contaminated portion. Salmonella spp. have the capability to enter a Viable but Non-Culturable (VBNC) state under unfavourable conditions, which adds to the challenges of culturing using various conventional culture media and enrichment protocols proposed for isolating Salmonella spp. from food and environmental samples (Pui et al., 2011).
-Morphological and biochemical characteristics
Subcultured isolates were examined for Gram's reaction and motility. Isolates of S. aureus were identified as Gram-positive cocci occurring singly or in diplo or staphylo arrangements. B. cereus isolates were confirmed as positive, rod-shaped bacteria occurring in short and long chains. Meanwhile, E. coli isolates exhibited Gram-negative, rod-shaped characteristics occurring singly or in diplo as observed under the microscope. In the motility test, S. aureus was found to be negative. All strains of B. cereus and E. coli demonstrated a diffuse cloud of growth away from the line of inoculation, indicating that all the strains were motile.
-IMViC test
Table 3 demonstrates the biochemical characteristics of the isolated bacteria. IMViC test showed that all S. aureus and B. cereus isolates tested negative for indole production and citrate utilisation. S. aureus isolates were positive for Methyl Red (MR) while B. cereus showed variable results. Variable results were also observed among S. aureus and B. cereus for Voges Proskauer (VP). Indole production and MR were positive for E. coli isolates, while VP and citrate utilisation were negative.
Table 3: Biochemical characterization of the isolates obtained from Vada pav samples
IMViC*=Indole, Methyl Red, Voges-Proskauer and Citrate test; TSI**=Triple Sugar Iron agar test; MR=Methyl Red; VP=Voges Proskauer; G=Gas; K=Alkaline; A=Acid; O=no colour change; NA=Not Applicable
E1 and E2=East zone; W1=West zone; N1=North zone; S1=South zone; C1 and C2=Central zone; a and b=trial
| Isolate | IMViC* | Enzymatic test | Sugar fermentation | TSI** agar | ||||||||||||||||||||||
| Indole | MR | VP | Citrate | Catalase | Oxidase | Urease | Nitratase | Gelatinase | Coagulase | Caseinase | Amylase | Hemolysis | Glucose | Sucrose | Lactose | Mannose | Arabinose | Mannitol | Dulcitol | Slant | Butt | H2S | Gas | Probable bacteria | ||
| C2a | - | + | + | - | + | - | + | + | - | - | NA | NA | - | + | + | + | + | + | + | + | A | A | - | - | Staphylococcus aureus | |
| E2a | - | + | - | - | + | - | + | - | + | - | NA | NA | - | + | + | + | + | + | + | + | A | A | - | - | ||
| E2b | - | + | - | - | + | - | + | - | + | - | NA | NA | - | + | + | + | + | + | + | + | A | O | - | - | ||
| S1b | - | - | + | - | + | - | + | + | + | NA | + | + | + | + | + | + | + | + | + | + | K | A | - | - | Bacillus cereus | |
| E2b | - | + | - | - | + | - | + | + | - | NA | + | + | + | + | + | + | + | + | + | + | A | A | - | - | ||
| C1b | + | + | - | - | + | - | - | + | - | NA | NA | NA | - | +G | +G | +G | +G | +G | +G | +G | A | A | - | + | Escherichia coli | |
| E1b | + | + | - | - | + | - | - | + | - | NA | NA | NA | - | +G | +G | +G | +G | +G | +G | +G | A | A | - | + | ||
| E2a | + | + | - | - | + | - | - | + | - | NA | NA | NA | - | +G | +G | +G | +G | +G | +G | +G | A | A | - | + | ||
E1 and E2=East zone; W1=West zone; N1=North zone; S1=South zone; C1 and C2=Central zone; a and b=trial
-Hemolysin production test
All B. cereus isolates were positive for hemolysin production; however, all S. aureus and E. coli isolates were found to be negative.
-Carbohydrate utilization test
S. aureus and B. cereus showed moderate to high carbohydrate utilization with no gas formation, while high utilization of carbohydrate with gas formation was observed in E. coli isolates.
-Triple sugar iron agar test
S. aureus isolates in general showed yellow-coloured slant and butts (indicating acid production) without gas or H2S production. Variable results were found in B. cereus isolates regarding slant colour showing either pink (alkaline) or yellow (acid) hue with acidic butt, no gas formation, and no H2S production. In contrast, E. coli isolates depicted an acidic slant and butt with gas formation and no H2S production.
-Enzyme production test
Most S. aureus isolates showed positive results for catalase, urea hydrolysis and gelatinase production, while they showed negative results for oxidase, nitratase, and coagulase tests. The majority of the B. cereus isolates were positive for all the enzyme tests except for the oxidase test, while E. coli isolates were negative for nearly all tests other than the catalase and nitratase tests.
Mandal and Mandal (2018) reported that S. aureus isolates were positive for MR, VP, citrate, catalase, gelatinase, and glucose utilization but negative for indole, oxidase, and urease. They noted variable results for nitratase production as well as sucrose, mannitol, and mannose utilization. The present study is consistent with these findings except for VP, urease, and gelatinase; furthermore, the isolates in this study utilized sucrose, mannitol, and mannose. Tewari et al. (2015) found that B. cereus isolates were motile and produced hemolysis while showing variable results for nitratase and VP tests; this study agrees with their findings. Bhutia et al. (2021) found that E. coli isolates were gram negative, motile, and gave positive results for arabinose utilization as well as catalase, nitratase, and MR tests while being negative for gelatinase, urease, and VP tests. Other tests such as indole, citrate, and carbohydrate utilization (glucose, lactose, sucrose, mannitol, and mannose) showed variable results. The present study showed similar results but E. coli strains were able to utilise almost all sugars with high acid and gas production.
Based on morphological and biochemical characterization, from a total of 14 samples tested: S. aureus (n=3), B. cereus (n=2), and E. coli (n=3) were identified.
Chumber et al. (2007) conducted the microbial assessment of street vended Vada pav samples and identified contamination with E. coli and Pseudomonas aeruginosa. Among the eight Vada pav samples tested, these bacterium was present in six samples. Similarly, in the present study, E. coli contamination was noted in three Vada pav samples from seven locations.
Severe contamination with S. aureus occurs due to improper handling, while the presence of E. coli and other coliform bacteria may result from insufficient hand washing and a lack of good manufacturing practices by food vendors (Mehta et al., 2020). B. cereus is extensively dispersed in natural environments and can be obtained from soil, water, and vegetation. The occurrence of Bacillus spp. isolates could result from the ability of Bacillus species to resist desiccation, allowing them to persist in dry products like grain and flour (Muhammad and Galadima, 2022).
Vendor location, raw material, utensils and equipment, storage and reheating, as well as the personal hygiene of the vendor are significant hazards and sources of microbial risks associated with contamination (Mohammed and Shehasen, 2020). According to Abdulkareem et al. (2014), vendors prepared food in unhygienic conditions, with flies present and stalls located near trash sites to prevent obstructions; these practices increase microbial hazards. Vendors often reuse water for hand washing, food preparation, and utensil cleaning, which can pose a risk to microbiological food safety (Prevolsek et al., 2021). The time and temperature of cooking are important as they may inactivate infectious bacteria that can develop during prolonged storage. Reheating temperatures must be sufficiently high or prolonged to effectively inactivate microbes. Some food vendors prepare food ahead of time and store it, then reheat the food at the time of sale. However, reheating the food is not always adequate for the destruction of microbes because the bacteria present in the food may have germinated from spores that survived cooking or multiplied after cooking (Rane, 2011).
Molecular characterization using PCR assay
The isolates were later subjected to PCR assay. Out of the isolates, two B. cereus isolatesand three E. coli isolates produced amplicons of the expected size as shown in Figure 2. Abdulrahman (2020) evaluated 200 whole chicken carcasses from Duhok, Kurdistan region, Iraq. Results showed that 28 out of 100 local chicken carcasses and 80 out of 100 imported frozen chicken carcasses were found to be contaminated with S. aureus using the conventional method. From the local chicken carcasses only 18 out of 22 coagulase-positive isolates were confirmed as S. aureus by amplification of the nuc gene; similary, from imported chicken carcasses 57 out of 68 coagulase-positive isolates were confirmed. The author concluded that PCR assay is more accurate and specific for the detection of S. aureus in food samples and that it seems to be more reliable than conventional methods for evaluating bacteriological safety of foods. In the present study out of three S. aureus isolates obtained using the conventional culture technique, none showed positive nuc gene amplification by PCR.
All B. cereus isolates were positive for hemolysin production; however, all S. aureus and E. coli isolates were found to be negative.
-Carbohydrate utilization test
S. aureus and B. cereus showed moderate to high carbohydrate utilization with no gas formation, while high utilization of carbohydrate with gas formation was observed in E. coli isolates.
-Triple sugar iron agar test
S. aureus isolates in general showed yellow-coloured slant and butts (indicating acid production) without gas or H2S production. Variable results were found in B. cereus isolates regarding slant colour showing either pink (alkaline) or yellow (acid) hue with acidic butt, no gas formation, and no H2S production. In contrast, E. coli isolates depicted an acidic slant and butt with gas formation and no H2S production.
-Enzyme production test
Most S. aureus isolates showed positive results for catalase, urea hydrolysis and gelatinase production, while they showed negative results for oxidase, nitratase, and coagulase tests. The majority of the B. cereus isolates were positive for all the enzyme tests except for the oxidase test, while E. coli isolates were negative for nearly all tests other than the catalase and nitratase tests.
Mandal and Mandal (2018) reported that S. aureus isolates were positive for MR, VP, citrate, catalase, gelatinase, and glucose utilization but negative for indole, oxidase, and urease. They noted variable results for nitratase production as well as sucrose, mannitol, and mannose utilization. The present study is consistent with these findings except for VP, urease, and gelatinase; furthermore, the isolates in this study utilized sucrose, mannitol, and mannose. Tewari et al. (2015) found that B. cereus isolates were motile and produced hemolysis while showing variable results for nitratase and VP tests; this study agrees with their findings. Bhutia et al. (2021) found that E. coli isolates were gram negative, motile, and gave positive results for arabinose utilization as well as catalase, nitratase, and MR tests while being negative for gelatinase, urease, and VP tests. Other tests such as indole, citrate, and carbohydrate utilization (glucose, lactose, sucrose, mannitol, and mannose) showed variable results. The present study showed similar results but E. coli strains were able to utilise almost all sugars with high acid and gas production.
Based on morphological and biochemical characterization, from a total of 14 samples tested: S. aureus (n=3), B. cereus (n=2), and E. coli (n=3) were identified.
Chumber et al. (2007) conducted the microbial assessment of street vended Vada pav samples and identified contamination with E. coli and Pseudomonas aeruginosa. Among the eight Vada pav samples tested, these bacterium was present in six samples. Similarly, in the present study, E. coli contamination was noted in three Vada pav samples from seven locations.
Severe contamination with S. aureus occurs due to improper handling, while the presence of E. coli and other coliform bacteria may result from insufficient hand washing and a lack of good manufacturing practices by food vendors (Mehta et al., 2020). B. cereus is extensively dispersed in natural environments and can be obtained from soil, water, and vegetation. The occurrence of Bacillus spp. isolates could result from the ability of Bacillus species to resist desiccation, allowing them to persist in dry products like grain and flour (Muhammad and Galadima, 2022).
Vendor location, raw material, utensils and equipment, storage and reheating, as well as the personal hygiene of the vendor are significant hazards and sources of microbial risks associated with contamination (Mohammed and Shehasen, 2020). According to Abdulkareem et al. (2014), vendors prepared food in unhygienic conditions, with flies present and stalls located near trash sites to prevent obstructions; these practices increase microbial hazards. Vendors often reuse water for hand washing, food preparation, and utensil cleaning, which can pose a risk to microbiological food safety (Prevolsek et al., 2021). The time and temperature of cooking are important as they may inactivate infectious bacteria that can develop during prolonged storage. Reheating temperatures must be sufficiently high or prolonged to effectively inactivate microbes. Some food vendors prepare food ahead of time and store it, then reheat the food at the time of sale. However, reheating the food is not always adequate for the destruction of microbes because the bacteria present in the food may have germinated from spores that survived cooking or multiplied after cooking (Rane, 2011).
Molecular characterization using PCR assay
The isolates were later subjected to PCR assay. Out of the isolates, two B. cereus isolatesand three E. coli isolates produced amplicons of the expected size as shown in Figure 2. Abdulrahman (2020) evaluated 200 whole chicken carcasses from Duhok, Kurdistan region, Iraq. Results showed that 28 out of 100 local chicken carcasses and 80 out of 100 imported frozen chicken carcasses were found to be contaminated with S. aureus using the conventional method. From the local chicken carcasses only 18 out of 22 coagulase-positive isolates were confirmed as S. aureus by amplification of the nuc gene; similary, from imported chicken carcasses 57 out of 68 coagulase-positive isolates were confirmed. The author concluded that PCR assay is more accurate and specific for the detection of S. aureus in food samples and that it seems to be more reliable than conventional methods for evaluating bacteriological safety of foods. In the present study out of three S. aureus isolates obtained using the conventional culture technique, none showed positive nuc gene amplification by PCR.
 |
.PNG) |
L=DNA Ladder (100bp), PC=Positive Control, NC=Negative Control, E 1 and E2=East 1 and East 2, S1=South 1, C1=Central 1, a and b=trial
In a study conducted by Tewari et al. (2015) on B. cereus contamination in raw meat and meat products, it was found that all B. cereus contained at least one of the four enterotoxin genes that encodingvirulence factors, 26 isolates out of a total 29 (89.7%) showed one gene from the NHE complex. The present study is comparable to this studies as all the isolates obtained (n = 2) were amplified for the nheA gene except for one.
In the study by Hegab et al. (2020), E. coli isolated using the conventional methods from Ras cheese (4/50), Domiati cheese (2/50) and Mish cheese (2/50) from Egypt also tested positive for the phoA gene in similar proportions. Eid et al. (2019) observed the presence of E. coli in minced meat, raw meat, sausage, burger, pastirma, luncheon, salami, and frankfurter using the conventional culture method; PCR results indicated that all E. coli isolates showed the presence of the phoA gene. The present study also showed similar findings where all E. coli isolates identified by the conventional culture method also amplified the phoA gene.
Table 4 shows the microbial contamination by location using both methods. Location E2 showed the presence of S. aureus, B. cereus, and E. coli, although S. aureus was not confirmed by the molecular method. Location E2 was an extremely congested spot close to heavy traffic and a railway line, which could explain the higher microbial contamination there. Overall, location E2 can be considered the lowest in hygienic practices while locations N1 and S1 were superior based on lower TVC values, absence of YMC, and all the other bacteria.
Table 5 shows a comparison between conventional and molecular methods for analysing microbial contamination in Vada pav samples.
In the study by Hegab et al. (2020), E. coli isolated using the conventional methods from Ras cheese (4/50), Domiati cheese (2/50) and Mish cheese (2/50) from Egypt also tested positive for the phoA gene in similar proportions. Eid et al. (2019) observed the presence of E. coli in minced meat, raw meat, sausage, burger, pastirma, luncheon, salami, and frankfurter using the conventional culture method; PCR results indicated that all E. coli isolates showed the presence of the phoA gene. The present study also showed similar findings where all E. coli isolates identified by the conventional culture method also amplified the phoA gene.
Table 4 shows the microbial contamination by location using both methods. Location E2 showed the presence of S. aureus, B. cereus, and E. coli, although S. aureus was not confirmed by the molecular method. Location E2 was an extremely congested spot close to heavy traffic and a railway line, which could explain the higher microbial contamination there. Overall, location E2 can be considered the lowest in hygienic practices while locations N1 and S1 were superior based on lower TVC values, absence of YMC, and all the other bacteria.
Table 5 shows a comparison between conventional and molecular methods for analysing microbial contamination in Vada pav samples.
Table 4: Microbial contamination of Vada pav samples by location
| Location | Trial | TVC log CFU/g |
YMC log CFU/g |
Staphylococcus aureus | Bacillus cereus | Escherichia coli |
| E1 | a | 5.86 | - | - | - | - |
| b | 6.31 | - | - | - | P | |
| E2 | a | 5.13 | 2.30 | P/N * | - | P |
| b | 5.08 | - | P/N * | P | - | |
| W1 | a | 3.48 | 4.28 | - | - | - |
| b | 4.39 | - | - | - | - | |
| N1 | a | 4.38 | - | - | - | - |
| b | 3.86 | - | - | - | - | |
| S1 | a | 4.13 | - | - | - | - |
| b | 4.58 | - | - | P | - | |
| C1 | a | 4.26 | 2.48 | - | - | - |
| b | 4.46 | 3.22 | - | - | P | |
| C2 | a | 3.87 | 2.70 | P/N * | - | - |
| b | 4.47 | - | - | - | - |
Table 5: Comparison of microbial conventional and molecular methods in Vada pav samples (n=7)
| Bacteria | Conventional culture technique | Molecular characterization | ||
| Positive examples (%) | Negative samples (%) | Positive examples (%) | Negative samples (%) | |
| Staphylococcus aureus | 2 (28.5%) | 5 (71.42%) | 0 | 7 (100%) |
| Bacillus cereus | 2 (28.5%) | 5 (71.42%) | 2 (28.5%) | 5 (71.42%) |
| Escherichia coli | 3 (42.85%) | 4 (57.14%) | 3 (42.85%) | 4 (57.14%) |
| Salmonella spp. | 0 | 7 (100%) | 0 | 7 (100%) |
Conclusion
Different locations significantly affected total viable count in Vada pav samples. The study also revealed contamination with S. aureus, B. cereus, and E. coli, with a higher incidence of E. coli (42.85%) compared to both B. cereus (28.57%) and S. aureus (28.57%). Salmonella spp. was not detected in any Vada pav samples. This study indicates a potential microbial health risk to humans through consumption of street vended Vada pav samples. Therefore, it is important to monitor and enhance hygienic practices among street food vendors.
Author contributions
M.D.P., P.V.H., and S.R. designed the study; M.D.P conducted the experimental work, analyzed the data, and wrote the manuscript. All authors reviewed and approved the final manuscript.
Conflicts of interest
The authors declare that there is no conflict of interest.
Acknowledgments
The authors express their gratitude to SHODH-Scheme of Developing High-Quality Research, Knowledge Consortium of Gujarat, Gujarat Education Department, Ahmedabad, Gujarat, India, for providing SHODH-fellowship.
The authors are extremely thankful to Dr. Prakash Koringa, Assistant Professor; Dr. Glory Parmar, Senior Research Fellow, Department of Animal Biotechnology; and Dr. Rafiyuddin Mathakiya, Assistant Professor, Department of Veterinary Microbiology, College of Veterinary Science and Animal Husbandry, Kamdhenu University, Anand 388001, Gujarat, India, for their valuable guidance in the molecular aspects of the research and for providing the facilities for conducting molecular characterization using PCR assay.
Funding
This study received no specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Ethical consideration
Not applicable.
References
Abdulkareem L., Garba D., Abubakar A. (2014). A study of cleanliness and sanitary practices of street food vendors in Northern Nigeria. Advances in Food Science and Technology. 2: 209-215.
Abdulrahman R.F. (2020). Detection of Staphylococcus aureus from local and imported chicken in Duhok province/Kurdistan region of Iraq using conventional and molecular methods. Basrah Journal of Veterinary Research. 19: 134-146.
Adhikari S., Sharma Regmi R., Sapkota S., Khadka S., Patel N., Gurung S., Thapa D., Bhattarai P., Sapkota P., Devkota R., Ghimire A., Rijal K.R. (2023). Multidrug resistance, biofilm formation and detection of blaCTX-M and blaVIM genes in E. coli and Salmonella isolates from chutney served at the street-food stalls of Bharatpur, Nepal. Heliyon. 9: e15739. [DOI: 10.1016/j.heliyon.2023.e15739].
Al-Ashmawy M.A., Sallam K.I., Abd-Elghany S.M., Elhadidy M., Tamura T. (2016). Prevalence, molecular characterization, and antimicrobial susceptibility of methicillin-resistant Staphylococcus aureus isolated from milk and dairy products. Foodborne Pathogens and Disease. 13: 156-162. [DOI: 10.1089/fpd.2015.2038]
Alelign D., Yihune M., Bekele M., Oumer Y., Beyene K., Atnafu K. (2023). Bacteriological quality and antimicrobial resistant patterns of foodborne pathogens isolated from commonly vended street foods in Arba Minch town, Southern Ethiopia. Infection and Drug Resistance. 16: 2883-2899. [DOI: 10.2147/ IDR.S411162]
Asiegbu C.V., Lebelo S.L., Tabit F.T. (2020). Microbial quality of ready-to-eat street vended food groups sold in the Johannesburg metropolis, South Africa. Journal of Food Quality and Hazards Control. 7: 18-26. [DOI: 10.18502/JFQHC.7.1.2448]
Bezerra A.C.D., Reis R.B.D., Bastos D.H.M. (2010). Microbiological quality of hamburgers sold in the streets of Cuiabá-MT, Brazil, and vendor hygiene-awareness. Food Science and Technology. 30: 520-524. [DOI:10.1590/S0101-20612010000200035]
Bhutia M.O., Thapa N., Tamang J.P. (2021). Prevalence of enterotoxin genes and antibacterial susceptibility pattern of pathogenic bacteria isolated from traditionally preserved fish products of Sikkim, India. Food Control. 125: 108009. [DOI: 10.1016/j.foodcont.2021.108009]
Braz V.S., Melchior K., Moreira C.G. (2020). Escherichia coli as a multifaceted pathogenic and versatile bacterium. Frontiers in Cellular and Infection Microbiology. 10: 548492. [DOI: 10.3389/fcimb.2020.548492]
Budiarso T.Y., Prihatmo G., Restiani R., Pakpahan S., Sari L. (2019). Detection of Staphylococcus aureus producing enterotoxin A on the skewers meatballs product in Yogyakarta City, Indonesia. Journal of Physics: Conference Series. 1397: 012044. [DOI: 10.1088/1742-6596/1397/1/012044]
Centre for Food Safety (CFS). (2014). Microbiological guidelines for food (for ready-to-eat food in general and specific food items). Food and Environmental Hygiene Department. Queensway, Hong Kong. URL: https://www.cfs.gov.hk/english/ food_leg/ files/food_leg_Microbiological_Guidelines_for_Food_e.pdf. Accessed 12 January 2024.
Chumber S.K., Kaushik K., Savy S. (2007). Bacteriological analysis of street foods in Pune. Editorial Board. 51: 83-136.
Eid H., El-Tabiy A., Fathy S. (2019). Molecular characterization of Escherichia coli isolated from meat and meat products in Port-Said markets. Suez Canal Veterinary Medical Journal. 24: 177-188. [DOI: 10.21608/SCVMJ.2019.69840]
Fisher E.L., Otto M., Cheung G.Y.C. (2018). Basis of virulence in enterotoxin-mediated staphylococcal food poisoning. Frontiers in Microbiology. 9: 436. [DOI: 10.3389/fmicb.2018.00436]
Foddai A.C., Grant I.R. (2020). Methods for detection of viable foodborne pathogens: Current state-of-art and future prospects. Applied Microbiology and Biotechnology. 104: 4281–4288. [DOI: 10.1007/s00253-020-10542-x]
Food Safety and Standards Authority of India (FSSAI). (2021). Guidelines for investigating and managing food-borne illness outbreaks in India. URL: https://fssai.gov.in/upload/advisories/2021/10/6177ebdb6d980Direction_Food_Bourne_Illness_26_10_2021.pdf. Accessed 22 February 2024.
Food Safety and Standards Authority of India (FSSAI). (2023). Microbiological standards of food grain products. URL: https://www.fssai.gov.in/upload/uploadfiles/files/20_%20Appendix%20B.pdf. Accessed 12 January 2024.
Food Standards Australia New Zealand (FSANZ). (2023). Agents of foodborne illness. Salmonella (non-typhoidal). Australian Government's Health, Canberra, Australia. URL: https://www.foodstandards.gov.au/publications/agents-foodborne-illness. Accessed 22 February 2024.
Hefny A., Mohamed H.M., Etokhy E.I., Abd El-Azeem M.W. (2020). Characterization of Bacillus cereus isolated from raw milk and milk products. Journal of Veterinary and Animal Research. 3: 205.
Hegab O.W., Abdel-Latif E.F., Moawad A.A. (2020). Isolation of enterotoxigenic Staphylococcus aureus harboring seb gene and enteropathogenic Escherichia coli (Serogroups O18, O114, and O125) from soft and hard artisanal cheeses in Egypt. Open Veterinary Journal. 10: 297-307. [DOI: 10.4314/ovj.v10i3.8]
Hirani D.R. (2019). Bacteriological analysis of street vended food panipuri in Mumbai Metropolitan Region. International Journal of Current Microbiology and Applied Sciences. 8: 115-121. [DOI: 10.20546/ijcmas.2019.811.014]
Hu Q., Tu J., Han X., Zhu Y., Ding C., Yu S. (2011). Development of multiplex PCR assay for rapid detection of Riemerella anatipestifer, Escherichia coli and Salmonella enterica simultaneously from ducks. Journal of Microbiological Methods. 87: 64-69. [DOI: 10.1016/j.mimet.2011.07.007]
Ibal J.C., Pham H.Q., Park C.E., Shin J.H. (2019). Information about variations in multiple copies of bacterial 16S rRNA genes may aid in species identification. PLoS One. 14: e0212090. [DOI: 10.1371/journal.pone.0212090]
Jotangiya D. (2018). Microbial assessment of hotdog–A popular street food of India and its comparison with homemade food. International Journal of Applied Home Science. 5: 343-346.
Kim J.H., Jung S., Oh S.W. (2020). Combination of bacteria concentration and DNA concentration for rapid detection of E. coli O157, L. monocytogenes, and S. Typhimurium without microbial enrichment. LWT. 117: 108609. [DOI: 10.1016/j.lwt.2019.108609]
Kumar T.D.K., Murali H.S., Batra H.V. (2009). Simultaneous detection of pathogenic B. cereus, S. aureus, and L. monocytogenes by multiplex PCR. Indian Journal of Microbiology. 49: 283-289. [DOI: 10.1007/s12088-009-0032-y]
Likhitha P., Nayak J.B., Thakur S. (2022). Prevalence of Staphylococcus aureus and methicillin-resistant Staphylococcus aureus in retail buffalo meat in Anand, India. The Pharma Innovation. 11: 17-20.
Liu C., Yu P., Yu S., Wang J., Guo H., Zhang Y., Zhang J., Liao X., Li C., Wu S., Gu Q., Zeng H., et al. (2020). Assessment and molecular characterization of Bacillus cereus isolated from edible fungi in China. BMC Microbiology. 20: 310. [DOI: 10.1186/s12866-020-01996-0]
Luo S., Liao C., Peng J., Tao S., Zhang T., Dai Y., Ding Y., Ma Y. (2023). Resistance and virulence gene analysis and molecular typing of Escherichia coli from duck farms in Zhanjiang, China. Frontiers in Cellular and Infection Microbiology. 13: 1202013. [DOI: 10.3389/fcimb.2023.1202013]
Mandal S., Mandal S. (2018). Multiple antibiotic resistance indices of potential pathogenic bacteria isolated from street vended fruit and sugarcane juices, Malda Town, India. Acta Scientific Pharmaceutical Sciences. 2:89-96.
Mehta H.D., Saradava D.A., Mehta D.N (2020). Bacteriological analysis and hygiene of street food panipuri: A case study of Morbi City-Gujarat, India. Indian Journal of Pure and Applied Biosciences. 8: 313-317. [DOI: 10.18782/2582-2845.8224]
Mohammed A.S., Shehasen M.Z. (2020). Street food consumption and associated health risk. International Journal of Research Studies in Agricultural Sciences. 6: 8-18. [DOI: 10.20431/2454-6224.0607002]
Muhammad Muhammad S., Ibrahim Galadima S. (2022). Determination of microbiological quality of bread and sanitation conditions of local bakeries in Aliero Town, Kebbi State. Applied Science and Technology Research Journal. 1: 1-9. [DOI: 10.31316/astro.v1i2.4274]
Nyabundi D., Onkoba N., Kimathi R., Nyachieo A., Juma G., Kinyanjui P., Kamau J. (2017). Molecular characterization and antibiotic resistance profiles of Salmonella isolated from fecal matter of domestic animals and animal products in Nairobi. Tropical Diseases, Travel Medicine and Vaccines. 3:2. [DOI: 10.1186/s40794-016-0045-6]
Patel R.J., Patel K.R. (2016). Experimental Microbiology. Aditya, India.
Prevolsek V., Ovca A., Jevšnik M. (2021). Fulfillment of technical and hygienic requirements among street food vendors in Slovenia. British Food Journal. 123: 105-123. [DOI: 10.1108/BFJ-11-2020-1056]
Pui C.F., Wong W.C., Chai L.C., Nillian E., Ghazali F.M., Cheah Y.K., Radu S. (2011). Simultaneous detection of Salmonella spp., Salmonella Typhi and Salmonella Typhimurium in sliced fruits using multiplex PCR. Food Control. 22: 337-342. [DOI: 10.1016/j.foodcont.2010.05.021]
Rajabi Z., M onadi Sefidan A., Zarebavani M., Sharifi Yazdi.S., Torabi Bonab P., Mirbagheri S.Z., Soltan-Dallal M.M. (2023). Investigation of enterotoxin-producing genes (sea, seb, sec, and sed) in Staphylococcus aureus isolated from raw traditionally and pasteurized milk supplied in Tehran, Iran. Journal of Food Quality and Hazards Control. 10: 221-225. [DOI: 10.18502/jfqhc.10.4.14180]
Rane S. (2011). Street vended food in developing world: Hazard analyses. Indian Journal of Microbiology. 51: 100-106. [DOI: 10.1007/s12088-011-0154-x]
Sharma D., Modgil R., Sandal A. (2020). Total viable microbial count of the selected street foods obtained from Palampur, India. Journal of Food Safety and Hygiene. 6: 47-52. [DOI: 10.18502/jfsh.v6i1.6026]
Sheth M., Gurudasani R., Mudbidri R. (2005). Screening for pathogenic microorganisms in street-vended bhelpuri in urban Vadodara: a HACCP approach. Journal of Food Science and Technology (Mysore). 42: 395-399.
Solanki D.G., Dave N.R. (2012). Nutritional and hygienic assessment of pizza sold by small vendors in Rajkot city and its comparison with homemade sample. Asian Journal of Home Science. 7: 31–34.
Solomon H. (2015). “The taste no chef can give”: Processing street food in Mumbai. Cultural Anthropology. 30: 65-90. [DOI: 10.14506/ca30.1.05]
Tewari A., Singh S.P., Singh R. (2015). Incidence and enterotoxigenic profile of Bacillus cereus in meat and meat products of Uttarakhand, India. Journal of Food Science and Technology. 52: 1796-1801. [DOI: 10.1007/s13197-013-1162-0]
Thakur S., Brahmbhatt M., Chaudhary J., Parmar B., Mistry U., Bhong C. (2020). Comparison of loop-mediated isothermal amplification with polymerase chain reaction for detection of methicillin-resistant Staphylococcus aureus in chevon. Journal of Entomology and Zoology Studies. 8: 1976-1980. [DOI: 10.22271/j.ento.2020.v8.i6aa.8111]
United Kingdom Health Security Agency (UKHSA). (2024), Guidelines for assessing the microbiological safety of ready-to-eat foods placed on the market. URL: https://assets.publishing. service.gov.uk/media/66debd72e87ad2f1218265e1/UKHSA-ready-to-eat-guidelines-2024.pdf. Accessed 17 January 2024.
World Health Organization (WHO). (2022). WHO global strategy for food safety 2022-2030: towards stronger food safety systems and global cooperation. World Health Organization. https://www. who.int/news-room/fact-sheets/detail/food-safety. Accessed 22 February 2024.
Yogesh M., Venkat Reddy D., Lahari Reddy K., Sri Mahitha G., Krishnaiah N. (2019). Studies on the microbiological quality of burgers sold in and around Greater Hyderabad Municipal Corporation. The Pharma Innovation Journal. 8: 264–268.
Zhao L., Wang J., Chen M., Sun X., Wang Y., Wang J., Geng Y. (2022). Development and application of recombinase polymerase amplification assays for rapid detection of Escherichia coli O157 in food. Food Analytical Methods. 15: 1843-1850. [DOI: 10.1007/s12161-022-02250-1]
Different locations significantly affected total viable count in Vada pav samples. The study also revealed contamination with S. aureus, B. cereus, and E. coli, with a higher incidence of E. coli (42.85%) compared to both B. cereus (28.57%) and S. aureus (28.57%). Salmonella spp. was not detected in any Vada pav samples. This study indicates a potential microbial health risk to humans through consumption of street vended Vada pav samples. Therefore, it is important to monitor and enhance hygienic practices among street food vendors.
Author contributions
M.D.P., P.V.H., and S.R. designed the study; M.D.P conducted the experimental work, analyzed the data, and wrote the manuscript. All authors reviewed and approved the final manuscript.
Conflicts of interest
The authors declare that there is no conflict of interest.
Acknowledgments
The authors express their gratitude to SHODH-Scheme of Developing High-Quality Research, Knowledge Consortium of Gujarat, Gujarat Education Department, Ahmedabad, Gujarat, India, for providing SHODH-fellowship.
The authors are extremely thankful to Dr. Prakash Koringa, Assistant Professor; Dr. Glory Parmar, Senior Research Fellow, Department of Animal Biotechnology; and Dr. Rafiyuddin Mathakiya, Assistant Professor, Department of Veterinary Microbiology, College of Veterinary Science and Animal Husbandry, Kamdhenu University, Anand 388001, Gujarat, India, for their valuable guidance in the molecular aspects of the research and for providing the facilities for conducting molecular characterization using PCR assay.
Funding
This study received no specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Ethical consideration
Not applicable.
References
Abdulkareem L., Garba D., Abubakar A. (2014). A study of cleanliness and sanitary practices of street food vendors in Northern Nigeria. Advances in Food Science and Technology. 2: 209-215.
Abdulrahman R.F. (2020). Detection of Staphylococcus aureus from local and imported chicken in Duhok province/Kurdistan region of Iraq using conventional and molecular methods. Basrah Journal of Veterinary Research. 19: 134-146.
Adhikari S., Sharma Regmi R., Sapkota S., Khadka S., Patel N., Gurung S., Thapa D., Bhattarai P., Sapkota P., Devkota R., Ghimire A., Rijal K.R. (2023). Multidrug resistance, biofilm formation and detection of blaCTX-M and blaVIM genes in E. coli and Salmonella isolates from chutney served at the street-food stalls of Bharatpur, Nepal. Heliyon. 9: e15739. [DOI: 10.1016/j.heliyon.2023.e15739].
Al-Ashmawy M.A., Sallam K.I., Abd-Elghany S.M., Elhadidy M., Tamura T. (2016). Prevalence, molecular characterization, and antimicrobial susceptibility of methicillin-resistant Staphylococcus aureus isolated from milk and dairy products. Foodborne Pathogens and Disease. 13: 156-162. [DOI: 10.1089/fpd.2015.2038]
Alelign D., Yihune M., Bekele M., Oumer Y., Beyene K., Atnafu K. (2023). Bacteriological quality and antimicrobial resistant patterns of foodborne pathogens isolated from commonly vended street foods in Arba Minch town, Southern Ethiopia. Infection and Drug Resistance. 16: 2883-2899. [DOI: 10.2147/ IDR.S411162]
Asiegbu C.V., Lebelo S.L., Tabit F.T. (2020). Microbial quality of ready-to-eat street vended food groups sold in the Johannesburg metropolis, South Africa. Journal of Food Quality and Hazards Control. 7: 18-26. [DOI: 10.18502/JFQHC.7.1.2448]
Bezerra A.C.D., Reis R.B.D., Bastos D.H.M. (2010). Microbiological quality of hamburgers sold in the streets of Cuiabá-MT, Brazil, and vendor hygiene-awareness. Food Science and Technology. 30: 520-524. [DOI:10.1590/S0101-20612010000200035]
Bhutia M.O., Thapa N., Tamang J.P. (2021). Prevalence of enterotoxin genes and antibacterial susceptibility pattern of pathogenic bacteria isolated from traditionally preserved fish products of Sikkim, India. Food Control. 125: 108009. [DOI: 10.1016/j.foodcont.2021.108009]
Braz V.S., Melchior K., Moreira C.G. (2020). Escherichia coli as a multifaceted pathogenic and versatile bacterium. Frontiers in Cellular and Infection Microbiology. 10: 548492. [DOI: 10.3389/fcimb.2020.548492]
Budiarso T.Y., Prihatmo G., Restiani R., Pakpahan S., Sari L. (2019). Detection of Staphylococcus aureus producing enterotoxin A on the skewers meatballs product in Yogyakarta City, Indonesia. Journal of Physics: Conference Series. 1397: 012044. [DOI: 10.1088/1742-6596/1397/1/012044]
Centre for Food Safety (CFS). (2014). Microbiological guidelines for food (for ready-to-eat food in general and specific food items). Food and Environmental Hygiene Department. Queensway, Hong Kong. URL: https://www.cfs.gov.hk/english/ food_leg/ files/food_leg_Microbiological_Guidelines_for_Food_e.pdf. Accessed 12 January 2024.
Chumber S.K., Kaushik K., Savy S. (2007). Bacteriological analysis of street foods in Pune. Editorial Board. 51: 83-136.
Eid H., El-Tabiy A., Fathy S. (2019). Molecular characterization of Escherichia coli isolated from meat and meat products in Port-Said markets. Suez Canal Veterinary Medical Journal. 24: 177-188. [DOI: 10.21608/SCVMJ.2019.69840]
Fisher E.L., Otto M., Cheung G.Y.C. (2018). Basis of virulence in enterotoxin-mediated staphylococcal food poisoning. Frontiers in Microbiology. 9: 436. [DOI: 10.3389/fmicb.2018.00436]
Foddai A.C., Grant I.R. (2020). Methods for detection of viable foodborne pathogens: Current state-of-art and future prospects. Applied Microbiology and Biotechnology. 104: 4281–4288. [DOI: 10.1007/s00253-020-10542-x]
Food Safety and Standards Authority of India (FSSAI). (2021). Guidelines for investigating and managing food-borne illness outbreaks in India. URL: https://fssai.gov.in/upload/advisories/2021/10/6177ebdb6d980Direction_Food_Bourne_Illness_26_10_2021.pdf. Accessed 22 February 2024.
Food Safety and Standards Authority of India (FSSAI). (2023). Microbiological standards of food grain products. URL: https://www.fssai.gov.in/upload/uploadfiles/files/20_%20Appendix%20B.pdf. Accessed 12 January 2024.
Food Standards Australia New Zealand (FSANZ). (2023). Agents of foodborne illness. Salmonella (non-typhoidal). Australian Government's Health, Canberra, Australia. URL: https://www.foodstandards.gov.au/publications/agents-foodborne-illness. Accessed 22 February 2024.
Hefny A., Mohamed H.M., Etokhy E.I., Abd El-Azeem M.W. (2020). Characterization of Bacillus cereus isolated from raw milk and milk products. Journal of Veterinary and Animal Research. 3: 205.
Hegab O.W., Abdel-Latif E.F., Moawad A.A. (2020). Isolation of enterotoxigenic Staphylococcus aureus harboring seb gene and enteropathogenic Escherichia coli (Serogroups O18, O114, and O125) from soft and hard artisanal cheeses in Egypt. Open Veterinary Journal. 10: 297-307. [DOI: 10.4314/ovj.v10i3.8]
Hirani D.R. (2019). Bacteriological analysis of street vended food panipuri in Mumbai Metropolitan Region. International Journal of Current Microbiology and Applied Sciences. 8: 115-121. [DOI: 10.20546/ijcmas.2019.811.014]
Hu Q., Tu J., Han X., Zhu Y., Ding C., Yu S. (2011). Development of multiplex PCR assay for rapid detection of Riemerella anatipestifer, Escherichia coli and Salmonella enterica simultaneously from ducks. Journal of Microbiological Methods. 87: 64-69. [DOI: 10.1016/j.mimet.2011.07.007]
Ibal J.C., Pham H.Q., Park C.E., Shin J.H. (2019). Information about variations in multiple copies of bacterial 16S rRNA genes may aid in species identification. PLoS One. 14: e0212090. [DOI: 10.1371/journal.pone.0212090]
Jotangiya D. (2018). Microbial assessment of hotdog–A popular street food of India and its comparison with homemade food. International Journal of Applied Home Science. 5: 343-346.
Kim J.H., Jung S., Oh S.W. (2020). Combination of bacteria concentration and DNA concentration for rapid detection of E. coli O157, L. monocytogenes, and S. Typhimurium without microbial enrichment. LWT. 117: 108609. [DOI: 10.1016/j.lwt.2019.108609]
Kumar T.D.K., Murali H.S., Batra H.V. (2009). Simultaneous detection of pathogenic B. cereus, S. aureus, and L. monocytogenes by multiplex PCR. Indian Journal of Microbiology. 49: 283-289. [DOI: 10.1007/s12088-009-0032-y]
Likhitha P., Nayak J.B., Thakur S. (2022). Prevalence of Staphylococcus aureus and methicillin-resistant Staphylococcus aureus in retail buffalo meat in Anand, India. The Pharma Innovation. 11: 17-20.
Liu C., Yu P., Yu S., Wang J., Guo H., Zhang Y., Zhang J., Liao X., Li C., Wu S., Gu Q., Zeng H., et al. (2020). Assessment and molecular characterization of Bacillus cereus isolated from edible fungi in China. BMC Microbiology. 20: 310. [DOI: 10.1186/s12866-020-01996-0]
Luo S., Liao C., Peng J., Tao S., Zhang T., Dai Y., Ding Y., Ma Y. (2023). Resistance and virulence gene analysis and molecular typing of Escherichia coli from duck farms in Zhanjiang, China. Frontiers in Cellular and Infection Microbiology. 13: 1202013. [DOI: 10.3389/fcimb.2023.1202013]
Mandal S., Mandal S. (2018). Multiple antibiotic resistance indices of potential pathogenic bacteria isolated from street vended fruit and sugarcane juices, Malda Town, India. Acta Scientific Pharmaceutical Sciences. 2:89-96.
Mehta H.D., Saradava D.A., Mehta D.N (2020). Bacteriological analysis and hygiene of street food panipuri: A case study of Morbi City-Gujarat, India. Indian Journal of Pure and Applied Biosciences. 8: 313-317. [DOI: 10.18782/2582-2845.8224]
Mohammed A.S., Shehasen M.Z. (2020). Street food consumption and associated health risk. International Journal of Research Studies in Agricultural Sciences. 6: 8-18. [DOI: 10.20431/2454-6224.0607002]
Muhammad Muhammad S., Ibrahim Galadima S. (2022). Determination of microbiological quality of bread and sanitation conditions of local bakeries in Aliero Town, Kebbi State. Applied Science and Technology Research Journal. 1: 1-9. [DOI: 10.31316/astro.v1i2.4274]
Nyabundi D., Onkoba N., Kimathi R., Nyachieo A., Juma G., Kinyanjui P., Kamau J. (2017). Molecular characterization and antibiotic resistance profiles of Salmonella isolated from fecal matter of domestic animals and animal products in Nairobi. Tropical Diseases, Travel Medicine and Vaccines. 3:2. [DOI: 10.1186/s40794-016-0045-6]
Patel R.J., Patel K.R. (2016). Experimental Microbiology. Aditya, India.
Prevolsek V., Ovca A., Jevšnik M. (2021). Fulfillment of technical and hygienic requirements among street food vendors in Slovenia. British Food Journal. 123: 105-123. [DOI: 10.1108/BFJ-11-2020-1056]
Pui C.F., Wong W.C., Chai L.C., Nillian E., Ghazali F.M., Cheah Y.K., Radu S. (2011). Simultaneous detection of Salmonella spp., Salmonella Typhi and Salmonella Typhimurium in sliced fruits using multiplex PCR. Food Control. 22: 337-342. [DOI: 10.1016/j.foodcont.2010.05.021]
Rajabi Z., M onadi Sefidan A., Zarebavani M., Sharifi Yazdi.S., Torabi Bonab P., Mirbagheri S.Z., Soltan-Dallal M.M. (2023). Investigation of enterotoxin-producing genes (sea, seb, sec, and sed) in Staphylococcus aureus isolated from raw traditionally and pasteurized milk supplied in Tehran, Iran. Journal of Food Quality and Hazards Control. 10: 221-225. [DOI: 10.18502/jfqhc.10.4.14180]
Rane S. (2011). Street vended food in developing world: Hazard analyses. Indian Journal of Microbiology. 51: 100-106. [DOI: 10.1007/s12088-011-0154-x]
Sharma D., Modgil R., Sandal A. (2020). Total viable microbial count of the selected street foods obtained from Palampur, India. Journal of Food Safety and Hygiene. 6: 47-52. [DOI: 10.18502/jfsh.v6i1.6026]
Sheth M., Gurudasani R., Mudbidri R. (2005). Screening for pathogenic microorganisms in street-vended bhelpuri in urban Vadodara: a HACCP approach. Journal of Food Science and Technology (Mysore). 42: 395-399.
Solanki D.G., Dave N.R. (2012). Nutritional and hygienic assessment of pizza sold by small vendors in Rajkot city and its comparison with homemade sample. Asian Journal of Home Science. 7: 31–34.
Solomon H. (2015). “The taste no chef can give”: Processing street food in Mumbai. Cultural Anthropology. 30: 65-90. [DOI: 10.14506/ca30.1.05]
Tewari A., Singh S.P., Singh R. (2015). Incidence and enterotoxigenic profile of Bacillus cereus in meat and meat products of Uttarakhand, India. Journal of Food Science and Technology. 52: 1796-1801. [DOI: 10.1007/s13197-013-1162-0]
Thakur S., Brahmbhatt M., Chaudhary J., Parmar B., Mistry U., Bhong C. (2020). Comparison of loop-mediated isothermal amplification with polymerase chain reaction for detection of methicillin-resistant Staphylococcus aureus in chevon. Journal of Entomology and Zoology Studies. 8: 1976-1980. [DOI: 10.22271/j.ento.2020.v8.i6aa.8111]
United Kingdom Health Security Agency (UKHSA). (2024), Guidelines for assessing the microbiological safety of ready-to-eat foods placed on the market. URL: https://assets.publishing. service.gov.uk/media/66debd72e87ad2f1218265e1/UKHSA-ready-to-eat-guidelines-2024.pdf. Accessed 17 January 2024.
World Health Organization (WHO). (2022). WHO global strategy for food safety 2022-2030: towards stronger food safety systems and global cooperation. World Health Organization. https://www. who.int/news-room/fact-sheets/detail/food-safety. Accessed 22 February 2024.
Yogesh M., Venkat Reddy D., Lahari Reddy K., Sri Mahitha G., Krishnaiah N. (2019). Studies on the microbiological quality of burgers sold in and around Greater Hyderabad Municipal Corporation. The Pharma Innovation Journal. 8: 264–268.
Zhao L., Wang J., Chen M., Sun X., Wang Y., Wang J., Geng Y. (2022). Development and application of recombinase polymerase amplification assays for rapid detection of Escherichia coli O157 in food. Food Analytical Methods. 15: 1843-1850. [DOI: 10.1007/s12161-022-02250-1]
* Corresponding author (T. Zeinali)
* E-mail: patelvh2004@yahoo.co.in
ORCID ID: https://orcid.org/0000-0003-4080-7899
* E-mail: patelvh2004@yahoo.co.in
ORCID ID: https://orcid.org/0000-0003-4080-7899
Type of Study: Original article |
Subject:
Special
Received: 24/03/31 | Accepted: 24/11/10 | Published: 24/12/30
Received: 24/03/31 | Accepted: 24/11/10 | Published: 24/12/30
References
1. Abdulkareem L., Garba D., Abubakar A. (2014). A study of cleanliness and sanitary practices of street food vendors in Northern Nigeria. Advances in Food Science and Technology. 2: 209-215.
2. Abdulrahman R.F. (2020). Detection of Staphylococcus aureus from local and imported chicken in Duhok province/Kurdistan region of Iraq using conventional and molecular methods. Basrah Journal of Veterinary Research. 19: 134-146. [DOI:10.23975/bjvetr.2020.170618]
3. Adhikari S., Sharma Regmi R., Sapkota S., Khadka S., Patel N., Gurung S., Thapa D., Bhattarai P., Sapkota P., Devkota R., Ghimire A., Rijal K.R. (2023). Multidrug resistance, biofilm formation and detection of blaCTX-M and blaVIM genes in E. coli and Salmonella isolates from chutney served at the street-food stalls of Bharatpur, Nepal. Heliyon. 9: e15739. [DOI: 10.1016/j.heliyon.2023.e15739]. [DOI:10.1016/j.heliyon.2023.e15739] [PMID] [PMCID]
4. Al-Ashmawy M.A., Sallam K.I., Abd-Elghany S.M., Elhadidy M., Tamura T. (2016). Prevalence, molecular characterization, and antimicrobial susceptibility of methicillin-resistant Staphylococcus aureus isolated from milk and dairy products. Foodborne Pathogens and Disease. 13: 156-162. [DOI: 10.1089/fpd.2015.2038] [DOI:10.1089/fpd.2015.2038] [PMID]
5. Alelign D., Yihune M., Bekele M., Oumer Y., Beyene K., Atnafu K. (2023). Bacteriological quality and antimicrobial resistant patterns of foodborne pathogens isolated from commonly vended street foods in Arba Minch town, Southern Ethiopia. Infection and Drug Resistance. 16: 2883-2899. [DOI: 10.2147/ IDR.S411162] [DOI:10.2147/IDR.S411162] [PMID] [PMCID]
6. Asiegbu C.V., Lebelo S.L., Tabit F.T. (2020). Microbial quality of ready-to-eat street vended food groups sold in the Johannesburg metropolis, South Africa. Journal of Food Quality and Hazards Control. 7: 18-26. [DOI: 10.18502/JFQHC.7.1.2448] [DOI:10.18502/jfqhc.7.1.2448]
7. Bezerra A.C.D., Reis R.B.D., Bastos D.H.M. (2010). Microbiological quality of hamburgers sold in the streets of Cuiabá-MT, Brazil, and vendor hygiene-awareness. Food Science and Technology. 30: 520-524. [DOI:10.1590/S0101-20612010000200035] [DOI:10.1590/S0101-20612010000200035]
8. Bhutia M.O., Thapa N., Tamang J.P. (2021). Prevalence of enterotoxin genes and antibacterial susceptibility pattern of pathogenic bacteria isolated from traditionally preserved fish products of Sikkim, India. Food Control. 125: 108009. [DOI: 10.1016/j.foodcont.2021.108009] [DOI:10.1016/j.foodcont.2021.108009]
9. Braz V.S., Melchior K., Moreira C.G. (2020). Escherichia coli as a multifaceted pathogenic and versatile bacterium. Frontiers in Cellular and Infection Microbiology. 10: 548492. [DOI: 10.3389/fcimb.2020.548492] [DOI:10.3389/fcimb.2020.548492] [PMID] [PMCID]
10. Budiarso T.Y., Prihatmo G., Restiani R., Pakpahan S., Sari L. (2019). Detection of Staphylococcus aureus producing enterotoxin A on the skewers meatballs product in Yogyakarta City, Indonesia. Journal of Physics: Conference Series. 1397: 012044. [DOI: 10.1088/1742-6596/1397/1/012044] [DOI:10.1088/1742-6596/1397/1/012044]
11. Centre for Food Safety (CFS). (2014). Microbiological guidelines for food (for ready-to-eat food in general and specific food items). Food and Environmental Hygiene Department. Queensway, Hong Kong. URL: https://www.cfs.gov.hk/english/ food_leg/ files/food_leg_Microbiological_Guidelines_for_Food_e.pdf. Accessed 12 January 2024.
12. Chumber S.K., Kaushik K., Savy S. (2007). Bacteriological analysis of street foods in Pune. Editorial Board. 51: 83-136.
13. Eid H., El-Tabiy A., Fathy S. (2019). Molecular characterization of Escherichia coli isolated from meat and meat products in Port-Said markets. Suez Canal Veterinary Medical Journal. 24: 177-188. [DOI: 10.21608/SCVMJ.2019.69840] [DOI:10.21608/scvmj.2019.69840]
14. Fisher E.L., Otto M., Cheung G.Y.C. (2018). Basis of virulence in enterotoxin-mediated staphylococcal food poisoning. Frontiers in Microbiology. 9: 436. [DOI: 10.3389/fmicb.2018.00436] [DOI:10.3389/fmicb.2018.00436] [PMID] [PMCID]
15. Foddai A.C., Grant I.R. (2020). Methods for detection of viable foodborne pathogens: Current state-of-art and future prospects. Applied Microbiology and Biotechnology. 104: 4281-4288. [DOI: 10.1007/s00253-020-10542-x] [DOI:10.1007/s00253-020-10542-x] [PMID] [PMCID]
16. Food Safety and Standards Authority of India (FSSAI). (2021). Guidelines for investigating and managing food-borne illness outbreaks in India. URL: https://fssai.gov.in/upload/advisories/2021/10/6177ebdb6d980Direction_Food_Bourne_Illness_26_10_2021.pdf. Accessed 22 February 2024.
17. Food Safety and Standards Authority of India (FSSAI). (2023). Microbiological standards of food grain products. URL: https://www.fssai.gov.in/upload/uploadfiles/files/20_%20Appendix%20B.pdf. Accessed 12 January 2024.
18. Food Standards Australia New Zealand (FSANZ). (2023). Agents of foodborne illness. Salmonella (non-typhoidal). Australian Government's Health, Canberra, Australia. URL: https://www.foodstandards.gov.au/publications/agents-foodborne-illness. Accessed 22 February 2024.
19. Hefny A., Mohamed H.M., Etokhy E.I., Abd El-Azeem M.W. (2020). Characterization of Bacillus cereus isolated from raw milk and milk products. Journal of Veterinary and Animal Research. 3: 205.
20. Hegab O.W., Abdel-Latif E.F., Moawad A.A. (2020). Isolation of enterotoxigenic Staphylococcus aureus harboring seb gene and enteropathogenic Escherichia coli (Serogroups O18, O114, and O125) from soft and hard artisanal cheeses in Egypt. Open Veterinary Journal. 10: 297-307. [DOI: 10.4314/ovj.v10i3.8] [DOI:10.4314/ovj.v10i3.8] [PMID] [PMCID]
21. Hirani D.R. (2019). Bacteriological analysis of street vended food panipuri in Mumbai Metropolitan Region. International Journal of Current Microbiology and Applied Sciences. 8: 115-121. [DOI: 10.20546/ijcmas.2019.811.014] [DOI:10.20546/ijcmas.2019.811.014]
22. Hu Q., Tu J., Han X., Zhu Y., Ding C., Yu S. (2011). Development of multiplex PCR assay for rapid detection of Riemerella anatipestifer, Escherichia coli and Salmonella enterica simultaneously from ducks. Journal of Microbiological Methods. 87: 64-69. [DOI: 10.1016/j.mimet.2011.07.007] [DOI:10.1016/j.mimet.2011.07.007] [PMID]
23. Ibal J.C., Pham H.Q., Park C.E., Shin J.H. (2019). Information about variations in multiple copies of bacterial 16S rRNA genes may aid in species identification. PLoS One. 14: e0212090. [DOI: 10.1371/journal.pone.0212090] [DOI:10.1371/journal.pone.0212090] [PMID] [PMCID]
24. Jotangiya D. (2018). Microbial assessment of hotdog-A popular street food of India and its comparison with homemade food. International Journal of Applied Home Science. 5: 343-346.
25. Kim J.H., Jung S., Oh S.W. (2020). Combination of bacteria concentration and DNA concentration for rapid detection of E. coli O157, L. monocytogenes, and S. Typhimurium without microbial enrichment. LWT. 117: 108609. [DOI: 10.1016/j.lwt.2019.108609] [DOI:10.1016/j.lwt.2019.108609]
26. Kumar T.D.K., Murali H.S., Batra H.V. (2009). Simultaneous detection of pathogenic B. cereus, S. aureus, and L. monocytogenes by multiplex PCR. Indian Journal of Microbiology. 49: 283-289. [DOI: 10.1007/s12088-009-0032-y] [DOI:10.1007/s12088-009-0032-y] [PMID] [PMCID]
27. Likhitha P., Nayak J.B., Thakur S. (2022). Prevalence of Staphylococcus aureus and methicillin-resistant Staphylococcus aureus in retail buffalo meat in Anand, India. The Pharma Innovation. 11: 17-20.
28. Liu C., Yu P., Yu S., Wang J., Guo H., Zhang Y., Zhang J., Liao X., Li C., Wu S., Gu Q., Zeng H., et al. (2020). Assessment and molecular characterization of Bacillus cereus isolated from edible fungi in China. BMC Microbiology. 20: 310. [DOI: 10.1186/s12866-020-01996-0] [DOI:10.1186/s12866-020-01996-0] [PMID] [PMCID]
29. Luo S., Liao C., Peng J., Tao S., Zhang T., Dai Y., Ding Y., Ma Y. (2023). Resistance and virulence gene analysis and molecular typing of Escherichia coli from duck farms in Zhanjiang, China. Frontiers in Cellular and Infection Microbiology. 13: 1202013. [DOI: 10.3389/fcimb.2023.1202013] [DOI:10.3389/fcimb.2023.1202013] [PMID] [PMCID]
30. Mandal S., Mandal S. (2018). Multiple antibiotic resistance indices of potential pathogenic bacteria isolated from street vended fruit and sugarcane juices, Malda Town, India. Acta Scientific Pharmaceutical Sciences. 2:89-96.
31. Mehta H.D., Saradava D.A., Mehta D.N (2020). Bacteriological analysis and hygiene of street food panipuri: A case study of Morbi City-Gujarat, India. Indian Journal of Pure and Applied Biosciences. 8: 313-317. [DOI: 10.18782/2582-2845.8224] [DOI:10.18782/2582-2845.8224]
32. Mohammed A.S., Shehasen M.Z. (2020). Street food consumption and associated health risk. International Journal of Research Studies in Agricultural Sciences. 6: 8-18. [DOI: 10.20431/2454-6224.0607002] [DOI:10.20431/2454-6224.0607002]
33. Muhammad Muhammad S., Ibrahim Galadima S. (2022). Determination of microbiological quality of bread and sanitation conditions of local bakeries in Aliero Town, Kebbi State. Applied Science and Technology Research Journal. 1: 1-9. [DOI: 10.31316/astro.v1i2.4274] [DOI:10.31316/astro.v1i2.4274]
34. Nyabundi D., Onkoba N., Kimathi R., Nyachieo A., Juma G., Kinyanjui P., Kamau J. (2017). Molecular characterization and antibiotic resistance profiles of Salmonella isolated from fecal matter of domestic animals and animal products in Nairobi. Tropical Diseases, Travel Medicine and Vaccines. 3:2. [DOI: 10.1186/s40794-016-0045-6] [DOI:10.1186/s40794-016-0045-6] [PMID] [PMCID]
35. Patel R.J., Patel K.R. (2016). Experimental Microbiology. Aditya, India.
36. Prevolsek V., Ovca A., Jevšnik M. (2021). Fulfillment of technical and hygienic requirements among street food vendors in Slovenia. British Food Journal. 123: 105-123. [DOI: 10.1108/BFJ-11-2020-1056] [DOI:10.1108/BFJ-11-2020-1056]
37. Pui C.F., Wong W.C., Chai L.C., Nillian E., Ghazali F.M., Cheah Y.K., Radu S. (2011). Simultaneous detection of Salmonella spp., Salmonella Typhi and Salmonella Typhimurium in sliced fruits using multiplex PCR. Food Control. 22: 337-342. [DOI: 10.1016/j.foodcont.2010.05.021] [DOI:10.1016/j.foodcont.2010.05.021]
38. Rajabi Z., M onadi Sefidan A., Zarebavani M., Sharifi Yazdi.S., Torabi Bonab P., Mirbagheri S.Z., Soltan-Dallal M.M. (2023). Investigation of enterotoxin-producing genes (sea, seb, sec, and sed) in Staphylococcus aureus isolated from raw traditionally and pasteurized milk supplied in Tehran, Iran. Journal of Food Quality and Hazards Control. 10: 221-225. [DOI: 10.18502/jfqhc.10.4.14180] [DOI:10.18502/jfqhc.10.4.14180]
39. Rane S. (2011). Street vended food in developing world: Hazard analyses. Indian Journal of Microbiology. 51: 100-106. [DOI: 10.1007/s12088-011-0154-x] [DOI:10.1007/s12088-011-0154-x] [PMID] [PMCID]
40. Sharma D., Modgil R., Sandal A. (2020). Total viable microbial count of the selected street foods obtained from Palampur, India. Journal of Food Safety and Hygiene. 6: 47-52. [DOI: 10.18502/jfsh.v6i1.6026] [DOI:10.18502/jfsh.v6i1.6026]
41. Sheth M., Gurudasani R., Mudbidri R. (2005). Screening for pathogenic microorganisms in street-vended bhelpuri in urban Vadodara: a HACCP approach. Journal of Food Science and Technology (Mysore). 42: 395-399.
42. Solanki D.G., Dave N.R. (2012). Nutritional and hygienic assessment of pizza sold by small vendors in Rajkot city and its comparison with homemade sample. Asian Journal of Home Science. 7: 31-34.
43. Solomon H. (2015). "The taste no chef can give": Processing street food in Mumbai. Cultural Anthropology. 30: 65-90. [DOI: 10.14506/ca30.1.05] [DOI:10.14506/ca30.1.05]
44. Tewari A., Singh S.P., Singh R. (2015). Incidence and enterotoxigenic profile of Bacillus cereus in meat and meat products of Uttarakhand, India. Journal of Food Science and Technology. 52: 1796-1801. [DOI: 10.1007/s13197-013-1162-0] [DOI:10.1007/s13197-013-1162-0] [PMID] [PMCID]
45. Thakur S., Brahmbhatt M., Chaudhary J., Parmar B., Mistry U., Bhong C. (2020). Comparison of loop-mediated isothermal amplification with polymerase chain reaction for detection of methicillin-resistant Staphylococcus aureus in chevon. Journal of Entomology and Zoology Studies. 8: 1976-1980. [DOI: 10.22271/j.ento.2020.v8.i6aa.8111] [DOI:10.22271/j.ento.2020.v8.i6aa.8111]
46. United Kingdom Health Security Agency (UKHSA). (2024), Guidelines for assessing the microbiological safety of ready-to-eat foods placed on the market. URL: https://assets.publishing. service.gov.uk/media/66debd72e87ad2f1218265e1/UKHSA-ready-to-eat-guidelines-2024.pdf. Accessed 17 January 2024.
47. World Health Organization (WHO). (2022). WHO global strategy for food safety 2022-2030: towards stronger food safety systems and global cooperation. World Health Organization. https://www. who.int/news-room/fact-sheets/detail/food-safety. Accessed 22 February 2024.
48. Yogesh M., Venkat Reddy D., Lahari Reddy K., Sri Mahitha G., Krishnaiah N. (2019). Studies on the microbiological quality of burgers sold in and around Greater Hyderabad Municipal Corporation. The Pharma Innovation Journal. 8: 264-268.
49. Zhao L., Wang J., Chen M., Sun X., Wang Y., Wang J., Geng Y. (2022). Development and application of recombinase polymerase amplification assays for rapid detection of Escherichia coli O157 in food. Food Analytical Methods. 15: 1843-1850. [DOI: 10.1007/s12161-022-02250-1] [DOI:10.1007/s12161-022-02250-1]
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |