Volume 12, Issue 4 (December 2025)
J. Food Qual. Hazards Control 2025, 12(4): 243-262 |
Back to browse issues page
Ethics code: Not applicable
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Jani J, Azizi Suhaili M, Lin C, Mandrinos S. Integrating Mitochondrial 12S rRNA Sequencing and Bioinformatics for Halal Authentication: A Review. J. Food Qual. Hazards Control 2025; 12 (4) :243-262
URL: http://jfqhc.ssu.ac.ir/article-1-1358-en.html
URL: http://jfqhc.ssu.ac.ir/article-1-1358-en.html
Department of Biomedical Sciences, Faculty of Medicine and Health Sciences, Universiti Malaysia Sabah, 88400 Kota Kinabalu Sabah, Malaysia, Borneo Medical and Health Research Centre, Faculty of Medicine and Health Sciences, Universiti Malaysia Sabah, 88400 Kota Kinabalu Sabah, Malaysia , jaeyres@ums.edu.my
Full-Text [PDF 962 kb]
(120 Downloads)
| Abstract (HTML) (90 Views)
Full-Text: (11 Views)
Integrating Mitochondrial 12S rRNA Sequencing and Bioinformatics for Halal Authentication: A Review
J. Jani 1,2[*]* , M. Azizi Suhaili 3, C.L.S. Lin 4, S. Mandrinos 5
1. Department of Biomedical Sciences, Faculty of Medicine and Health Sciences, Universiti Malaysia Sabah, 88400 Kota Kinabalu Sabah, Malaysia
2. Borneo Medical and Health Research Centre, Faculty of Medicine and Health Sciences, Universiti Malaysia Sabah, 88400 Kota Kinabalu Sabah, Malaysia
3. Department of Public Health Medicine, Faculty of Medicine and Health Sciences, Universiti Malaysia Sabah, 88400 Kota Kinabalu Sabah, Malaysia
4. Department of Anesthesiology and Intensive Care, Faculty of Medicine and Health Science, 88400 Kota Kinabalu Sabah, Malaysia
5. School of Business Swinburne, University of Technology, 93350 Kuching, Sarawak, Malaysia
HIGHLIGHTS
Table 1: Several justifications highlighting the effectiveness of the 12S rRNA gene from mitochondrial DNA in the context of halal verification
rRNA=Ribosomal RNA
Table 2: Advantages and Disadvantages of PacBio sequencing platform
Table 5: Polymerase chain reaction species forward and reverse (5’ to 3’) products
Table 6: Bioinformatics tools for metagenomics analysis in halal authentication
J. Jani 1,2[*]*
1. Department of Biomedical Sciences, Faculty of Medicine and Health Sciences, Universiti Malaysia Sabah, 88400 Kota Kinabalu Sabah, Malaysia
2. Borneo Medical and Health Research Centre, Faculty of Medicine and Health Sciences, Universiti Malaysia Sabah, 88400 Kota Kinabalu Sabah, Malaysia
3. Department of Public Health Medicine, Faculty of Medicine and Health Sciences, Universiti Malaysia Sabah, 88400 Kota Kinabalu Sabah, Malaysia
4. Department of Anesthesiology and Intensive Care, Faculty of Medicine and Health Science, 88400 Kota Kinabalu Sabah, Malaysia
5. School of Business Swinburne, University of Technology, 93350 Kuching, Sarawak, Malaysia
- The mitochondrial 12S ribosomal RNA gene served as a robust molecular marker for precise species identification in halal authentication.
- Integration of next generation sequencing and third generation sequencing enhanced the accuracy and scalability of halal verification.
- This framework promoted transparency, sustainability, and consumer trust in global halal supply chains.
| Article type Review article |
ABSTRACT This study proposed a molecular framework to ensure compliance with Islamic dietary laws, aligning with the rapidly expanding global halal market projected to reach USD 3 trillion by 2027. The approach integrated mitochondrial 12S ribosomal RNA sequencing with bioinformatics tools using Next-Generation Sequencing (NGS) and Third-Generation Sequencing (TGS) technologies to facilitate accurate species identification, DNA barcoding, and meet origin authentication. The 12S ribosomal RNA gene provides high precision for species verification due to its conserved yet variable sequence regions. A bioinformatics pipeline incorporating Mothur and QIIME2 was developed to analyse sequencing outputs and validate species Operational Taxonomic Units (OTUs). This pipeline enhanced traceability and ensured compliance with halal regulatory standards. By detecting even trace residues of non-halal substances, the method strengthens food safety, prevents cross-contamination, and increases consumer trust. The significance of these molecular techniques is in ensuring the safety of food and preventing spoilage. By detecting residues of non-halal substances, this approach addressed significant concerns related to cross-contamination and labelling inaccuracies, enhancing transparency and strengthening consumer trust. Analysing halal certification practices as per industry traditions and international regulatory bodies, as well as the thought-provoking shift to align high-end technologies and foreign experts requires careful harmonisation. This study also focused on factors contributing to that harmonisation. To assess the possible future of halal verification, this review presented a prospective, amalgamating modern science with religious justice to achieve a functional balance and preserve the reliability of halal-validated products. These innovations enable higher levels of precision, transparency, and sustainability, which foster consumer trust and reinvigorate the global marketplace. © 2025, Shahid Sadoughi University of Medical Sciences. This is an open access article under the Creative Commons Attribution 4.0 International License. |
||
| Keywords Certification Mitochondria Bioinformatics Sequence Analysis DNA |
|||
| Article history Received: 20 Apr 2025 Revised: 12 Nov 2025 Accepted: 25 Nov 2025 |
|||
| Abbreviations gDNA=genomic DNA mtDNA=mitochondrial DNA NGS=Next Generation Sequencing ONT=Oxford Nanopore Technologies PCR=Polymerase Chain Reaction PE=Paired End ROI=Region of Interest rRNA=ribosomal RNA TGS=Third Generation Sequencing |
To cite: Jani J., Azizi Suhaili M., Lin C.L.S., Mandrinos S. (2025). Integrating mitochondrial 12S rRNA sequencing and bioinformatics for halal authentication: a review. Journal of Food Quality and Hazards Control. 12: 243-262.
Introduction
Introduction
The Malaysian Investment Development Authority (MIDA, 2022) cited a projection that the global halal market will expand from USD 2 trillion in 2021 to USD 3 trillion (approximately RM 13.3 trillion) by 2027, based on a report by Business Wire. This remarkable growth is fuelled by the rising global Muslim population, which accounts for about 24% of the world’s population, or approximately 1.9 billion people as of 2022. Halal food products encompass several major categories, including meat, poultry, processed seafood, dairy products, cereals and grains, oils and fats, confectionery, and processed fruits and vegetables. The largest demand originates from Asia, followed by the Middle East, Africa, Europe, North America, and Latin America (Shaharuddin et al., 2025). With over 300 global food and beverage companies based in Asia, Malaysia has emerged as a leading hub for halal manufacturing and certification, earning widespread international trust and acceptance for its halal-compliant products (MIDA, 2022; Neequaye et al., 2017).
Malaysia’s halal ecosystem is spearheaded by the Jabatan Agama Islam Malaysia (JAKIM) and the Halal Development Corporation Berhad (HDC), which are responsible for regulatory oversight, certification, and policy development. These agencies align their operations with the Halal Industry Masterplan 2030 to strengthen the nation’s infrastructure and human capital in halal management (Nasir et al., 2021). However, despite Malaysia’s leadership in halal certification, one of the key challenges in maintaining halal authenticity lies in the integration of state-of-the-art technologies for verification and traceability (Hassan et al., 2020).
In this context, the convergence of molecular sequencing and halal verification offers a timely and transformative opportunity. The duality between traditional certification systems and cutting-edge genomic technologies creates both potential and paradoxes that require critical exploration (Garinet et al., 2018). As consumer awareness increases and global supply chains become more complex, ensuring transparency and reliability in halal authentication is no longer feasible through manual inspection and document-based verification alone. Traditional techniques, though valuable, are often time-consuming and inadequate in detecting cross-contamination or fraud in the modern food industry (Amer, 2024).
The emergence of bioinformatics has revolutionised biological data analysis, offering high-throughput, data-centric methods for authenticity verification. Bioinformatics provides computational pipelines capable of analysing large volumes of sequencing data, thereby enabling species identification, DNA barcoding, and halal status determination with unparalleled accuracy (Zou et al., 2023). Studies such as Shen et al. (2022) have demonstrated that bioinformatics-based halal integrity systems improve traceability, enhance food safety, and ensure data-driven decision-making across the supply chain. Applications include mitochondrial 12S ribosomal RNA (rRNA) sequencing for species identification, DNA-based quantification of halal components, and detection of non-halal residues in processed food.
Therefore, the purpose of this study was to develop and propose a comprehensive molecular and computational framework for halal authentication using mitochondrial 12S rRNA sequencing integrated with bioinformatics analysis pipelines such as Mothur and QIIME2. This approach aims to establish a scientifically robust, traceable, and high-throughput system capable of verifying species origin, preventing cross-contamination, and supporting regulatory compliance within the halal supply chain. By aligning genomic science with Islamic dietary law, this study seeks to bridge the gap between modern molecular technologies and traditional halal certification practices, ensuring transparency, sustainability, and consumer confidence in the global halal marketplace.
Ultimately, this study contributes to the future of halal verification by harmonising technological innovation with religious and industrial integrity, thereby positioning bioinformatics as a cornerstone of sustainable halal authentication and global trade assurance (Chen et al., 2023).
Mitochondria and the mitochondrial 12S rRNA gene
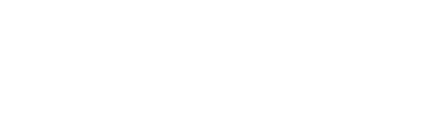
Mitochondria are double-membraned organelles often referred to as the powerhouses of the cell due to their critical role in energy production through oxidative phosphorylation. They possess their own genetic material, known as mitochondrial DNA (mtDNA), which is circular and distinct from the nuclear genome (Monzel et al., 2023). mtDNA encodes 13 essential proteins involved in the electron transport chain, along with 22 transfer RNAs (tRNAs) and two rRNAs, namely the 12S and 16S rRNA genes, which are vital for mitochondrial protein synthesis (Figure 1). The unique characteristics of mtDNA, including maternal inheritance, high copy number, lack of recombination, and relatively rapid mutation rate, make it a powerful molecular marker for evolutionary, taxonomic, and forensic studies (Ferreira and Rodriguez, 2024).
Structurally, the mitochondrion comprises an outer membrane that encloses the organelle and an inner membrane folded into cristae, which increases the surface area for ATP synthesis. The space between these membranes is called the intermembrane space, while the innermost compartment, the matrix, contains the mitochondrial genome, ribosomes, and enzymes essential for the citric acid cycle (Absmeier et al., 2023). This semi-autonomous nature allows mitochondria to regulate their own protein synthesis and maintain genetic continuity across generations.
Malaysia’s halal ecosystem is spearheaded by the Jabatan Agama Islam Malaysia (JAKIM) and the Halal Development Corporation Berhad (HDC), which are responsible for regulatory oversight, certification, and policy development. These agencies align their operations with the Halal Industry Masterplan 2030 to strengthen the nation’s infrastructure and human capital in halal management (Nasir et al., 2021). However, despite Malaysia’s leadership in halal certification, one of the key challenges in maintaining halal authenticity lies in the integration of state-of-the-art technologies for verification and traceability (Hassan et al., 2020).
In this context, the convergence of molecular sequencing and halal verification offers a timely and transformative opportunity. The duality between traditional certification systems and cutting-edge genomic technologies creates both potential and paradoxes that require critical exploration (Garinet et al., 2018). As consumer awareness increases and global supply chains become more complex, ensuring transparency and reliability in halal authentication is no longer feasible through manual inspection and document-based verification alone. Traditional techniques, though valuable, are often time-consuming and inadequate in detecting cross-contamination or fraud in the modern food industry (Amer, 2024).
The emergence of bioinformatics has revolutionised biological data analysis, offering high-throughput, data-centric methods for authenticity verification. Bioinformatics provides computational pipelines capable of analysing large volumes of sequencing data, thereby enabling species identification, DNA barcoding, and halal status determination with unparalleled accuracy (Zou et al., 2023). Studies such as Shen et al. (2022) have demonstrated that bioinformatics-based halal integrity systems improve traceability, enhance food safety, and ensure data-driven decision-making across the supply chain. Applications include mitochondrial 12S ribosomal RNA (rRNA) sequencing for species identification, DNA-based quantification of halal components, and detection of non-halal residues in processed food.
Therefore, the purpose of this study was to develop and propose a comprehensive molecular and computational framework for halal authentication using mitochondrial 12S rRNA sequencing integrated with bioinformatics analysis pipelines such as Mothur and QIIME2. This approach aims to establish a scientifically robust, traceable, and high-throughput system capable of verifying species origin, preventing cross-contamination, and supporting regulatory compliance within the halal supply chain. By aligning genomic science with Islamic dietary law, this study seeks to bridge the gap between modern molecular technologies and traditional halal certification practices, ensuring transparency, sustainability, and consumer confidence in the global halal marketplace.
Ultimately, this study contributes to the future of halal verification by harmonising technological innovation with religious and industrial integrity, thereby positioning bioinformatics as a cornerstone of sustainable halal authentication and global trade assurance (Chen et al., 2023).
Mitochondria and the mitochondrial 12S rRNA gene
Mitochondria are double-membraned organelles often referred to as the powerhouses of the cell due to their critical role in energy production through oxidative phosphorylation. They possess their own genetic material, known as mitochondrial DNA (mtDNA), which is circular and distinct from the nuclear genome (Monzel et al., 2023). mtDNA encodes 13 essential proteins involved in the electron transport chain, along with 22 transfer RNAs (tRNAs) and two rRNAs, namely the 12S and 16S rRNA genes, which are vital for mitochondrial protein synthesis (Figure 1). The unique characteristics of mtDNA, including maternal inheritance, high copy number, lack of recombination, and relatively rapid mutation rate, make it a powerful molecular marker for evolutionary, taxonomic, and forensic studies (Ferreira and Rodriguez, 2024).
Structurally, the mitochondrion comprises an outer membrane that encloses the organelle and an inner membrane folded into cristae, which increases the surface area for ATP synthesis. The space between these membranes is called the intermembrane space, while the innermost compartment, the matrix, contains the mitochondrial genome, ribosomes, and enzymes essential for the citric acid cycle (Absmeier et al., 2023). This semi-autonomous nature allows mitochondria to regulate their own protein synthesis and maintain genetic continuity across generations.

Figure 1: Overview of domestic animal species and mitochondrial DNA structure. Silhouettes depict representative livestock (goat, cattle, horse, sheep, dog, pig, and rat). The lower panels illustrate (left) the mitochondrion and its internal structure, (center) the circular mitochondrial genome (~16.5 kb) showing 12S and 16S rRNA regions, and (right) the secondary structure of 12S rRNA with protein-binding sites.
mDNA=Mitochondrial DNA; rRNA=Ribosomal RNA
mDNA=Mitochondrial DNA; rRNA=Ribosomal RNA
Researchers seldom utilize the mitochondrial 12S rRNA gene as a primary marker in molecular studies (Luo et al., 2011). In fact, the normal method of halal validation is based on DNA or protein fragments extracted from specific tissues, such as in the case of meat or food form itself, to ensure they meet Islamic dietary requirements (Ng et al., 2021). The 12S rRNA gene is widely used as a molecular marker for a variety of applications, such as DNA barcoding, phylogenetics, and species identification (Chan et al., 2022). Although DNA-based methods have simple yet useful applications in food authentication and traceability, including halal authentication, they often have limitations in specifically addressing the genetic markers of the species being studied or targeting the presence of forbidden phytochemicals (Muflihah et al., 2023).
With techniques like Polymerase Chain Reaction (PCR), DNA tracking, and real-time PCR, various genetic markers in halal products can be analysed, and authentication for halal status can be granted. These methods have been utilised effectively in determining the source of meat products by assessing some types of mtDNA (Murugaiah et al., 2009). Factors such as the researcher’s aims, and the type of food product being analysed impact on halal authentication technologies and markers (Lubis et al., 2016). Researchers and regulatory authorities typically adopt a combination of scientific approaches and traditional approaches in halal certification methods (Ng et al., 2021).
The 12S rRNA gene, derived from mtDNA, has been identified as an effective target for halal authentication, as shown in Table 1. The specificity of the mitochondrial genome favours species identification, due to maternal inheritance and a higher number of cellular copies than the nuclear DNA (Cahyadi et al., 2020).
With techniques like Polymerase Chain Reaction (PCR), DNA tracking, and real-time PCR, various genetic markers in halal products can be analysed, and authentication for halal status can be granted. These methods have been utilised effectively in determining the source of meat products by assessing some types of mtDNA (Murugaiah et al., 2009). Factors such as the researcher’s aims, and the type of food product being analysed impact on halal authentication technologies and markers (Lubis et al., 2016). Researchers and regulatory authorities typically adopt a combination of scientific approaches and traditional approaches in halal certification methods (Ng et al., 2021).
The 12S rRNA gene, derived from mtDNA, has been identified as an effective target for halal authentication, as shown in Table 1. The specificity of the mitochondrial genome favours species identification, due to maternal inheritance and a higher number of cellular copies than the nuclear DNA (Cahyadi et al., 2020).
Table 1: Several justifications highlighting the effectiveness of the 12S rRNA gene from mitochondrial DNA in the context of halal verification
| Justifications | Details | References |
| Species Identification | The 12S rRNA gene was examined using Next Generation Sequencing (NGS) technologies, enabling precise determination of the animal species from which the meat originates. This is crucial for ensuring that the meat is sourced from animals that are halal. | (Zhang et al., 2020) |
| DNA Barcoding | The 12S rRNA gene was used as a DNA barcode for Halal authentication. A reference library of verified 12S rRNA sequences was created from recognised halal species to enable the comparison and confirmation of sample tests. | (Cahyadi et al., 2020) |
| Sequence Variability | Examine variations in the 12S rRNA gene sequence to differentiate between species that are closely related. This can be particularly important in cases where different species might have similar morphological characteristics. | (Chan et al., 2022) |
| Quantitative Analysis |
Quantitative PCR (qPCR) or other methods were used to quantify the amount of mitochondrial DNA, specifically the 12S rRNA gene, in each sample collected. This quantitative approach can provide insights into the composition of meat mixtures. | (Rohman et al., 2022) |
| Authentication of Meat Origin | The authenticity of the declared meat origin was confirmed by comparing the obtained 12S rRNA sequences with reference sequences of known halal animals. This aids in detecting potential adulteration or labelling errors. | (Cahyadi et al., 2020) |
| High-Throughput Screening | The high-throughput capabilities of Next Generation Sequencing (NGS) and Third Generation Sequencing (TGS) were leveraged to process large numbers of samples simultaneously. This is particularly useful for screening meat products in industrial settings or at different points along the supply chain. | (Liu et al., 2021) |
| Validation with Regulatory Standards | The 12S rRNA gene was used to validate halal authentication against established regulatory standards and guidelines. Collaboration with relevant regulatory bodies is necessary to ensure acceptance and recognition of the method. | (Noor et al., 2023) |
| Integration into Existing Certification Processes | The analysis of the 12S rRNA gene was integrated into existing halal certification processes to enhance the accuracy and reliability of halal verification. This ensures alignment with industry practices and standards. | (Van et al., 2012) |
| Cross-validation with Other Molecular Markers | The 12S rRNA gene obtained was cross validated with other molecular markers or methods commonly used in halal certification to strengthen the overall authenticity testing approach. | (Galal-Khallaf, 2021) |
Molecular characterisation of halal authenticity has focused on representative profiles of the meat fraction, with a relative upper trifling ratio found through the latter target (the mitochondrial 12S rRNA gene). However, the mitochondrial target of interest has also been the subject of investigation in other studies. The availability of this molecular strategy for sensitive halal authentication in the food industries will be beneficial for researchers and stakeholders who wish to apply it across different thermal processing techniques (Kaveh et al., 2023; Rahman et al., 2023). In addition, the mitochondrial 12S rRNA gene serves as a unique molecular marker utilised in sequencing technologies, providing a high-throughput quantitative tool for species identification (Woo et al., 2023), DNA barcoding (Lücking et al., 2020), and meat authentication studies concerning halal certification and compliance with Islamic dietary laws (Aziz et al., 2023). This progress paves the way for a practical and science-based route to authenticating the genuineness and integrity of halal products within the food supply system (Jaswir et al., 2016).
Revolutionising halal verification with Next Generation Sequencing (NGS) and Third Generation Sequencing (TGS) technologies
In recent years, the emergence of high-throughput sequencing platforms such as and TGS has revolutionised the field of genomics. These technologies have enabled the rapid, large-scale acquisition of genetic data, thereby transforming research in areas such as biodiversity, medicine, agriculture, and food authentication. Companies including Illumina, Pacific Biosciences (PacBio), and Oxford Nanopore Technologies (ONT) have played a pivotal role in advancing these platforms, providing comprehensive tools for genome analysis and molecular characterisation across a wide range of species (Liu et al., 2023).
NGS represents a major advancement over conventional Sanger sequencing due to its ability to generate millions of short reads simultaneously, reducing both time and cost. This high-throughput capacity allows for extensive genetic profiling, species identification, and DNA barcoding at an unprecedented resolution. In the context of halal authentication, NGS facilitates the detection of trace DNA sequences from mixed or processed food products, which traditional methods might fail to identify. This makes NGS particularly valuable in verifying the presence or absence of non-halal components, ensuring the accuracy and reliability of halal certification (Aini et al., 2023).
The application of NGS in halal verification offers several advantages. First, it enables species-level identification through targeted sequencing of mitochondrial genes such as 12S rRNA, which are conserved across species but exhibit sufficient variability for precise differentiation. Second, it provides a digital, traceable record of genetic data, thereby increasing transparency and accountability throughout the halal supply chain. Third, when coupled with advanced bioinformatics tools like Mothur and QIIME2, NGS data can be processed, classified, and validated against comprehensive genetic databases to confirm species origin and authenticity with high confidence.
Beyond its technical merits, NGS plays an essential role in strengthening the integrity of halal verification systems by integrating scientific objectivity with religious compliance. By detecting even minute levels of DNA contamination, NGS safeguards against cross-contamination during food processing and enhances consumer trust in certified halal products. Moreover, as the halal market continues to expand globally, NGS provides the scalability and efficiency required to support industrial-scale testing and regulatory enforcement.
In addition to NGS, TGS technologies such as PacBio’s Single Molecule Real-Time (SMRT) sequencing and Oxford Nanopore’s nanopore-based platforms further extend analytical capabilities by producing long-read sequences that enable comprehensive genome assembly and structural variant detection. The integration of NGS and TGS technologies offers a hybrid approach, combining the accuracy of short reads with the continuity of long reads to achieve superior precision in species identification and genomic interpretation (Aini et al., 2023). Therefore, the adoption of NGS and TGS technologies in halal verification represents a paradigm shift from conventional biochemical testing towards a molecular, data-driven framework. This scientific evolution not only enhances the credibility and traceability of halal authentication but also aligns Malaysia’s halal certification system with global standards of food safety and technological innovation.
It is important to note that the field of sequencing technologies is constantly evolving, and new advancements may have emerged since the author’s recent update. Also, when selecting a sequencing platform, the specific needs and resources pertinent to the halal industry should be carefully considered.
PacBio sequencing
PacBio, which stands for Pacific Biosciences sequencing, is a TGS method recognised for its long-read capabilities (Rhoads and Au, 2015). This technology has both advantages and disadvantages aspects (Table 2) that are worth considering across various industries, including those related to halal products. The PacBio sequencing platform offers benefits such as high accuracy, extended read lengths, and the ability to characterise complex genomic regions. However, it is essential to consider the associated costs, limitations in throughput, and sample prerequisites in relation to the specific needs and available resources of the halal industry.
Revolutionising halal verification with Next Generation Sequencing (NGS) and Third Generation Sequencing (TGS) technologies
In recent years, the emergence of high-throughput sequencing platforms such as and TGS has revolutionised the field of genomics. These technologies have enabled the rapid, large-scale acquisition of genetic data, thereby transforming research in areas such as biodiversity, medicine, agriculture, and food authentication. Companies including Illumina, Pacific Biosciences (PacBio), and Oxford Nanopore Technologies (ONT) have played a pivotal role in advancing these platforms, providing comprehensive tools for genome analysis and molecular characterisation across a wide range of species (Liu et al., 2023).
NGS represents a major advancement over conventional Sanger sequencing due to its ability to generate millions of short reads simultaneously, reducing both time and cost. This high-throughput capacity allows for extensive genetic profiling, species identification, and DNA barcoding at an unprecedented resolution. In the context of halal authentication, NGS facilitates the detection of trace DNA sequences from mixed or processed food products, which traditional methods might fail to identify. This makes NGS particularly valuable in verifying the presence or absence of non-halal components, ensuring the accuracy and reliability of halal certification (Aini et al., 2023).
The application of NGS in halal verification offers several advantages. First, it enables species-level identification through targeted sequencing of mitochondrial genes such as 12S rRNA, which are conserved across species but exhibit sufficient variability for precise differentiation. Second, it provides a digital, traceable record of genetic data, thereby increasing transparency and accountability throughout the halal supply chain. Third, when coupled with advanced bioinformatics tools like Mothur and QIIME2, NGS data can be processed, classified, and validated against comprehensive genetic databases to confirm species origin and authenticity with high confidence.
Beyond its technical merits, NGS plays an essential role in strengthening the integrity of halal verification systems by integrating scientific objectivity with religious compliance. By detecting even minute levels of DNA contamination, NGS safeguards against cross-contamination during food processing and enhances consumer trust in certified halal products. Moreover, as the halal market continues to expand globally, NGS provides the scalability and efficiency required to support industrial-scale testing and regulatory enforcement.
In addition to NGS, TGS technologies such as PacBio’s Single Molecule Real-Time (SMRT) sequencing and Oxford Nanopore’s nanopore-based platforms further extend analytical capabilities by producing long-read sequences that enable comprehensive genome assembly and structural variant detection. The integration of NGS and TGS technologies offers a hybrid approach, combining the accuracy of short reads with the continuity of long reads to achieve superior precision in species identification and genomic interpretation (Aini et al., 2023). Therefore, the adoption of NGS and TGS technologies in halal verification represents a paradigm shift from conventional biochemical testing towards a molecular, data-driven framework. This scientific evolution not only enhances the credibility and traceability of halal authentication but also aligns Malaysia’s halal certification system with global standards of food safety and technological innovation.
It is important to note that the field of sequencing technologies is constantly evolving, and new advancements may have emerged since the author’s recent update. Also, when selecting a sequencing platform, the specific needs and resources pertinent to the halal industry should be carefully considered.
PacBio sequencing
PacBio, which stands for Pacific Biosciences sequencing, is a TGS method recognised for its long-read capabilities (Rhoads and Au, 2015). This technology has both advantages and disadvantages aspects (Table 2) that are worth considering across various industries, including those related to halal products. The PacBio sequencing platform offers benefits such as high accuracy, extended read lengths, and the ability to characterise complex genomic regions. However, it is essential to consider the associated costs, limitations in throughput, and sample prerequisites in relation to the specific needs and available resources of the halal industry.
Table 2: Advantages and Disadvantages of PacBio sequencing platform
| Advantages and Disadvantage |
Details | References |
| Accuracy in Genome Sequencing | PacBio sequencing provides high accuracy in genome sequencing, which is crucial for identifying and verifying the genetic makeup of organisms relevant to the halal industry. | (Rhoads and Au, 2015) |
| Long Read Lengths | PacBio sequencing produces long reads, enabling the sequencing of entire genes or even entire genomes without the need for assembly from short reads. This is beneficial for accurately identifying and characterising specific genes or regions of interest. | (Hook and Timp, 2023) |
| Detection of Genetic Variations | The long-read lengths facilitate the detection of genetic variations, such as Single Nucleotide Polymorphisms (SNPs) and structural variations. This is important in ensuring the authenticity and genetic integrity of halal products. | (Bannor et al., 2023; Sajali et al., 2020) |
| Characterisation of Complex Genomic Regions | PacBio sequencing is well-suited for characterising complex genomic regions, including repetitive elements and structural variations, which can be challenging for other sequencing technologies. This is valuable for a comprehensive understanding of genetic traits in halal-relevant organisms. | (Rhoads and Au, 2015) |
| Cost | PacBio sequencing is more expensive compared to other sequencing technologies. This can be a limiting factor, especially for large-scale genomic studies in the halal industry. | (Teng et al., 2017) |
| Throughput | While PacBio sequencing provides long reads, its throughput can be lower compared to other sequencing platforms. This limitation may impact the speed at which large-scale genomic data can be generated. | (Amarasinghe et al., 2020) |
| Error Rate | Although PacBio sequencing has made progress in accuracy, it may still show a greater incidence of errors when compared with some short-read sequencing technologies. This can be a concern when precise genetic information is crucial for halal certification. | (Oehler et al., 2023; Sedlazeck et al., 2018) |
| Sample Requirements | PacBio sequencing may have specific sample conditions, and the challenge of sourcing high-quality DNA from certain types of samples may restrict its applicability in the halal industry. | (De Coster et al., 2021; Haynes et al., 2019) |
ONT sequencing
The concept of “MinION sequencing” likely refers to sequencing performed using the MinION sequencer from ONT. ONT offers a portable DNA and RNA sequencing platform that utilises nanopore technology for real-time analysis. There are several advantages and disadvantages of ONT sequencing, as shown in Table 3.
The concept of “MinION sequencing” likely refers to sequencing performed using the MinION sequencer from ONT. ONT offers a portable DNA and RNA sequencing platform that utilises nanopore technology for real-time analysis. There are several advantages and disadvantages of ONT sequencing, as shown in Table 3.
Table 3: Advantages and disadvantages of ONT sequencing platform
ONT=Oxford Nanopore Technologies
| Advantages and Disadvantages | Details | References |
| Real-Time Sequencing | The ONT sequencing provides real-time data, allowing for rapid analysis and decision-making. This can be beneficial in the halal industry for quick verification of the genetic makeup of organisms or products. | (Tyler et al., 2018) |
| Portability | The ONT sequencer is portable and relatively small, making it suitable for field applications. This can be advantageous in situations where on-site sequencing is needed, such as in remote areas or at production facilities in the halal industry. | (Wasswa et al., 2022) |
| Long Read Lengths | Similar to PacBio sequencing, ONT sequencing also offers long read lengths. This is advantageous for accurately characterising genes and genomic regions relevant to the halal industry without the need for assembly. | (Udaondo et al., 2021) |
| Ease of Use | The ONT sequencer is known for its user-friendly interface and simplified workflow. This can be advantageous in settings where technical expertise may be limited, such as in small-scale halal production facilities. | (King et et al., 2020; Wasswa et al., 2022) |
| Cost-Effective for Small-Scale Projects | The ONT sequencer has relatively low upfront costs compared to some other sequencing technologies. This makes it potentially cost-effective for small-scale projects or applications in the halal industry with limited budgets. | (Chang et al., 2020; Wasswa et al., 2022) |
| Error Rates | Nanopore sequencing technologies, including ONT, have historically had higher error rates compared to some other sequencing platforms. While error rates have improved over time, accuracy may still be a concern, especially for applications requiring precise genetic information. | (Sun et al., 2022) |
| Throughput | While ONT sequencing is suitable for small-scale projects, it may have limitations in terms of throughput for large-scale genomic studies. High-throughput sequencing technologies may be more appropriate for large-scale applications in the halal industry. | (Haynes et al., 2019; Wasswa et al., 2022) |
| Sample Contamination | Nanopore sequencing is sensitive to sample quality, and contamination can affect the accuracy of results. Ensuring high-quality DNA is essential for reliable sequencing, which may be a consideration in the halal industry where sample quality can vary. | (Haynes et al., 2019; Noakes et al., 2019) |
| Data Storage and Analysis | The real-time nature of ONT sequencing generates large amounts of data, and handling and analysing this data may require advanced computational resources. This can be a consideration for facilities with limited bioinformatics capabilities in the halal industry. | (Cao et al., 2015; Lu et al., 2016) |
Illumina sequencing
Illumina sequencing represents a leading technology in NGS that has established itself as a standard in genomics research. Table 4 outlines some potential advantages and disadvantages of applying Illumina sequencing within the halal industry.
Given its high accuracy, particularly when combined with other applications, along with its high throughput and versatility, Illumina sequencing holds undeniable promise for many applications in the halal sector, particularly those that require fast and reliable analysis (Meyer and Kircher, 2010). Still, the limited read lengths, size of the instrument, and associated costs must all be weighed according to the specific requirements of genomic assays.
The ultimate decision is and should be application-based, with sequencing technology selected based on the unmet needs and the unique requirements of the halal industry. Considering that PacBio sequencing is capable of generating long reads, it is particularly useful for characterising complex genomic regions and variations (Karst et al., 2021) making it a valuable tool for complete genomic characterisation. Conversely, if the application involves real-time data analytics or requires mobility, ONT sequencing is a more compact and portable option. However, it is important to note that this approach has a lower output and a higher error rate. With its combination of accuracy, throughput, and cost, Illumina sequencing has become particularly beneficial, enabling a wide array of high-throughput applications and providing researchers with a wide versatility of applications to serve their needs.
Given the variety of options available, the prevailing view among researchers is that a hybrid approach, using multiple sequencing technologies can leverage the strengths of each platform for a more holistic analysis (Jovic et al., 2022). Nevertheless, the sequence data itself, along with financial and logistical planning for specific projects, will ultimately determine the best sequencing method to accommodate the specific needs of halal authenticity testing.
Illumina sequencing represents a leading technology in NGS that has established itself as a standard in genomics research. Table 4 outlines some potential advantages and disadvantages of applying Illumina sequencing within the halal industry.
Given its high accuracy, particularly when combined with other applications, along with its high throughput and versatility, Illumina sequencing holds undeniable promise for many applications in the halal sector, particularly those that require fast and reliable analysis (Meyer and Kircher, 2010). Still, the limited read lengths, size of the instrument, and associated costs must all be weighed according to the specific requirements of genomic assays.
The ultimate decision is and should be application-based, with sequencing technology selected based on the unmet needs and the unique requirements of the halal industry. Considering that PacBio sequencing is capable of generating long reads, it is particularly useful for characterising complex genomic regions and variations (Karst et al., 2021) making it a valuable tool for complete genomic characterisation. Conversely, if the application involves real-time data analytics or requires mobility, ONT sequencing is a more compact and portable option. However, it is important to note that this approach has a lower output and a higher error rate. With its combination of accuracy, throughput, and cost, Illumina sequencing has become particularly beneficial, enabling a wide array of high-throughput applications and providing researchers with a wide versatility of applications to serve their needs.
Given the variety of options available, the prevailing view among researchers is that a hybrid approach, using multiple sequencing technologies can leverage the strengths of each platform for a more holistic analysis (Jovic et al., 2022). Nevertheless, the sequence data itself, along with financial and logistical planning for specific projects, will ultimately determine the best sequencing method to accommodate the specific needs of halal authenticity testing.
Table 4: Advantages and disadvantages of Illumina sequencing platform
| Advantages and Disadvantages | Details | References |
| High Accuracy | Illumina sequencing is known for its high accuracy, which is crucial for obtaining reliable genetic information. This accuracy is important in halal industry applications, such as the identification and verification of specific genetic traits in organisms. | (Haynes et al., 2019; Muflihah et al., 2023) |
| High Throughput | The Illumina platforms typically offer high throughput, allowing for the simultaneous sequencing of multiple samples. This is advantageous for large-scale genomic studies in the halal industry, where the analysis of a significant number of samples may be required. | (Reuter et al., 2015) |
| Cost-Effective for High-Throughput Applications | Illumina sequencing is often considered cost-effective for high-throughput projects. The cost per base pair is relatively low, making it suitable for projects in the halal industry that require extensive genomic coverage. | (Satam et al., 2023) |
| Wide Range of Applications | Illumina sequencing platforms are versatile and can be used for various applications, including whole-genome sequencing, RNA sequencing, and targeted sequencing. This flexibility allows researchers in the halal industry to choose the most suitable approach for their specific needs. | (Haynes et al., 2019) |
| Established Protocols and Workflows | Illumina sequencing has been widely adopted in the scientific community, and there are well-established protocols and workflows. This can simplify the sequencing process and make it more accessible, even in settings with limited expertise in genomics. | (Satam et al., 2023) |
| Short Read Lengths | One limitation of Illumina sequencing is the relatively short read lengths compared to some other sequencing technologies. This may pose challenges when trying to sequence through repetitive or complex genomic regions in the halal industry. | (Bianconi et al., 2023) |
| Instrument Size | Some Illumina sequencers may be larger and less portable compared to certain portable sequencing technologies. If on-site or field sequencing is required in the halal industry, the size and portability of the sequencer could be a consideration. | (Guptaand Verma, 2019; Slatko et al., 2018) |
| Error Rate in Homopolymeric Regions | Illumina sequencing can have difficulties accurately sequencing homopolymeric regions (repeated sequences of the same nucleotide). This may be a concern in applications where these regions are relevant, such as in the analysis of certain genes in the halal industry. | (Moorthie et al., 2011) |
| Complex Library Preparation | The library preparation for Illumina sequencing can be more complex compared to some other sequencing platforms. This may add an extra step and potentially increase the chances of errors or contamination in the halal industry workflow. | (Yang et al., 2014) |
| Instrument Costs | While Illumina sequencing can be cost-effective for high-throughput applications, the upfront costs of purchasing the sequencing instruments can be relatively high. This may be a consideration for facilities with limited budgets in the halal industry. | (Dunham and Friesen, 2013) |
Design of specialised primers intended for the identification of the 12S rRNA gene
The PCR technique is typically carried out to selectively amplify the 12S rRNA genes from the extracted DNA. Several primers can be proposed for use in a multiplex setup of the mitochondrial 12S rRNA gene, as shown in Table 5. Within these primers, there exists a universal sequence that targets the Region of Interest (ROI).
The PCR technique is typically carried out to selectively amplify the 12S rRNA genes from the extracted DNA. Several primers can be proposed for use in a multiplex setup of the mitochondrial 12S rRNA gene, as shown in Table 5. Within these primers, there exists a universal sequence that targets the Region of Interest (ROI).
Table 5: Polymerase chain reaction species forward and reverse (5’ to 3’) products
| Animals | Sequences | Base pair (bp) | References |
| Goat | F 5’-GACCTCCCAGCTCCATCAAACATCTCATCTTGATGAAA-3’ R 5’-CTCGACAAATGTGAGTTACAGAGGGA-3’ |
157 | (Matsunaga et al., 1999) |
| Chicken | F 5’-GACCTCCCAGCTCCATCAAACATCTCATCTTGATGAAA-3’ R 5’-AAGATACAGATGAAGAAGAATGAGGCG-3’ |
227 | (Matsunaga et al., 1999) |
| F 5’-TTCTTCGGACACCCCGAAG-3’ R 5’-CTAGGCCCAGGAAATGTT-3’ |
596 | (Boyrusbianto et al., 2023) | |
| Cattle | F 5’-GACCTCCCAGCTCCATCAAACATCTCATCTTGATGAAA-3’ R 5’-CTAGAAAAGTGTAAGACCCGTAATATAAG-3’ |
274 | (Matsunaga et al., 1999) |
| Sheep | F 5’-GACCTCCCAGCTCCATCAAACATCTCATCTTGATGAAA-3’ R 5’-CTATGAATGCTGTGGCTATTGTCGCA-3’ |
331 | (Matsunaga et al., 1999) |
| Horse | F 5’-GACCTCCCAGCTCCATCAAACATCTCATCTTGATGAAA-3’ R 5’-CTCAGATTCACTCGACGAGGGTAGTA-3’ |
439 | (Matsunaga et al., 1999) |
| Rat | F 5’-GACCTCCCAGCTCCATCAAACATCTCATCTTGATGAAA-3’ R 5’-GAATGGGATTTTGTCTGCGTTGGAGTTT-3’ |
603 | (Matsunaga et al.,1999; Nuraini et al., 2012) |
| F 5’-ACCGCGGTCATACGATTAAC-3’ R 5’-TCTGGGAAAAGAAAATGTAGCC-3’ |
491 | (Cahyadi et al., 2020) | |
| Bovine | F 5’-ACCGCGGTCATACGATTAAC-3’ R 5’-AGTGCGTCGGCTATTGTAGG-3’ |
155 | (Cahyadi et al., 2020) |
| F 5’-TTCTTCGGACACCCCGAAG-3’ R 5’-CGGTTGGAATAGCAATAA-3’ |
263 | (Boyrusbianto et al., 2023) | |
| Dog | F 5’-ACCGCGGTCATACGATTAAC-3’ R 5’-TCCTCTGGCGAATTATTTTGTTG-3’ |
244 | (Cahyadi et al., 2020) |
| Pig | F 5’-ACCGCGGTCATACGATTAAC-3’ R 5’-GAATTGGCAAGGGTTGGTAA-3’ |
357 | (Cahyadi et al., 2020) |
| F 5’-TTCTTCGGACACCCCGAAG-3’ R 5’-TGGTGAGCCCATACGATA-3’ |
168 | (Boyrusbianto et al., 2023) | |
| F 5’-GACCTCCCAGCTCCATCAAACATCTCATCTTGATGAAA-3’ R 5’-CTGATAGTAGATTTGTGATGACCGTA-3’ |
398 |
(Matsunaga et al., 1999) | |
| F 5’-AACCCTATGTACGTCGTGCAT-3’ R 5’-ACCATTGACTGAATAGCACCT-3’ |
531 | (Monteil-Sosa et al., 2000; Sahilah et al 2011) | |
| Primers universal | F 5’- AAACTGGGATTAGATACCCCACTA -3’ R 5’-GAGGGTGACGGGCGGTGTGT-3’ |
456 | (Pandey et al., 2007; Sahilah et al., 2011) |
Table 5 summarises the forward and reverse primer sequences used for PCR-based species identification, specifically targeting various animal species relevant to halal authentication. The table lists the primers, expected PCR product sizes (in base pairs, bp), and corresponding references, demonstrating the diversity of genetic markers used across different studies.
The table includes primers for multiple commonly consumed animals such as goat, chicken, cattle, sheep, horse, rat, dog, and pig. These species are frequently tested in halal verification to ensure the absence of non-halal ingredients or cross-contamination in food products. In addition, universal primers are included, which target conserved regions of the mitochondrial 12S rRNA gene and can amplify DNA across multiple vertebrate species.
Each primer set is associated with an expected amplicon size, which ranges from 155 bp (bovine) to 603 bp (rat), depending on the species and the genetic region targeted. Shorter amplicons are often preferred for processed food products, where DNA may be partially degraded. Longer amplicons may provide higher specificity in differentiating closely related species. For instance, the chicken primers produce amplicons of 227 bp and 596 bp, allowing flexibility depending on the sample type and quality.
The type of sample analysed using these primers can include raw meat, processed food, mixed meat products, or highly processed ingredients, all of which are relevant in halal authentication. The number of samples tested in each study may vary, but PCR amplification with these primers provides a rapid, sensitive, and species-specific method for detecting halal or non-halal components. For example, primers designed for pig DNA (non-halal) are critical for identifying contamination in food products labelled as halal.
References in Table 5 highlight the original sources where these primers were developed or validated, such as Matsunaga et al. (1999), Cahyadi et al. (2020), and Boyrusbianto et al. (2023). These studies demonstrated the reliability and reproducibility of these primers for molecular authentication. By integrating these primers into a PCR or sequencing workflow, halal verification can be performed accurately and efficiently, enabling both regulatory compliance and consumer confidence.
Overall, Table 5 illustrates the comprehensive set of primers available for PCR-based species identification, supporting the use of mtDNA markers such as 12S rRNA for halal authentication across a wide range of food products.
Optimising PCR amplification for high-quality sequencing across Illumina, PacBio, and ONT platforms
PCR amplification is a major process to enhance sequencing outputs for platforms such as Illumina, PacBio, and ONT. This step measures that the target ROI, like the 12S rRNA gene from genomic DNA (gDNA) input, is present in the proper quantity and quality, as both are essential for fetching high and reliable sequencing results (Kayama et al., 2021). Specific primers can be used to improve the selective amplification of target sequence, concentrating sequencing activity in the ROI and achieving high coverage, even for low-abundance regions (Ye et al., 2012).
The first steps of such a workflow typically involve the use of DNA extraction kits and protocols that protect DNA integrity (Dubois et al., 2022). From here, high-quality gDNA is used as an input for PCR amplification (Schloss et al., 2011). Next, primers are designed for the target ROI. For example, in the case of the 12S rRNA gene, primers are strategically chosen to bind conserved sequences flanking the target region, thereby clinching specificity (Woo et al., 2023). On the Illumina platform, primers are designed to amplify shorter DNA fragments (ranging from 250 to 300 bp) to ensure compatibility with the minimum read length required for Paired End (PE) sequencing (Kozich et al., 2013). On the other hand, large amplicon sequencing technologies (hundreds to a few kilobases), such as PacBio and ONT use primers designed to amplify the large amplicons, taking advantage of their long-read capabilities (Liu et al., 2020).
The cycle of denaturation, annealing, and extension is then repeated to achieve exponential amplification of the target genetic sequence (Nichols and Quince, 2016). The number of cycles is optimised based on the starting gDNA quantity and the desired amplicon length. Then, the amplified PCR products are purified to remove unincorporated primers, nucleotides, and other contaminants that might adversely affect sequencing. With long-read technologies, size selection is also applicable to ensure that the amplicons are of optimal size for sequencing. Depending on the platform, the next step of library preparation differs. For Illumina sequencing, the putative amplicons are ligated with platform-specific adapters, which ultimately form a sequencing library to be sequenced via PE or single-end (SE) strategies (Callahan et al., 2016). In a different approach, the PacBio and ONT libraries apply the ligation of adapters onto the amplified amplicons to prime them for long-read sequencing. Long-read platforms excel at resolving challenging areas, such as repeat elements and whole-gene analysis, including the 12S rRNA.
Once the sequencing libraries are ready, data generation begins. The second approach employed by Illumina is high-throughput, short-read sequencing, which delivers highly accurate data but often needs overlapping PE reads to cover the entire area of interest (Callahan et al., 2016). PacBio and ONT, on the other hand, use long-read sequencing methods that allow them to capture entire regions in a single pass, leading to easier assembly and more accurate mapping of challenging genomic regions. While gDNA serves the same fundamental role across all sequencing platforms, differences exist in the specific amplification and sequencing processes used (Woltyńska et al., 2023). PCR amplification enriches the signal of the desired sequence to achieve sufficient flow-cell coverage necessary for quality sequencing. Optimising workflows for respective platforms maximises sequencing potential, particularly in regions such as the 12S rRNA gene (Cahyadi et al., 2020). This includes Illumina short-read sequencing technology, which provides high throughput over small DNA fragments (Callahan et al., 2016). Meanwhile, long-read sequencing through PacBio and ONT offers better identification of structural variants and improved resolution of complex genomic segments (Ahsan et al., 2023).
To summarise, PCR amplification is an enabling step in sequencing workflows, enabling technologies, such as Illumina, PacBio, and ONT for a variety of applications. Non-Long Terminal Repeat (non-LTR) retrotransposons could certainly interfere with PCR amplification and reduce sequencing yields, though PCR remains essential for enriching target regions as well as ensuring a reliable sequence of interest. By implementing a bespoke amplification process, sequencing platforms have expanded their applications in species identification, phylogenetics, and mitochondrial genome studies (Schloss et al., 2011).
Short-read versus long-read sequencing: a comprehensive approach to 12S rRNA analysis for bioinformatics
It is widely known that Illumina sequencing technology boasts high throughput and outstanding accuracy, thus making it an optimal platform for producing short reads to cover target sequences, even when they do not exceed 500 bp (Kozich et al., 2013). Two-read analysis on mitochondrial 12S rRNA using Illumina’s PE sequencing format for 250 PE or 300 PE reads has been performed (Prakrongrak et al., 2023). However, the short-read property of Illumina sequencing makes it difficult to merge overlaps from PE reads simply, hence it typically needs to be preceded by tools like FLASH, PEAR, or BBMerge. These tools perform synchronisation and merging of the complementary reads to reconstruct the sequence of the entire target in question while ensuring that everything is covered correctly with minimal or no gaps or assembly variants. Such methods improve downstream bioinformatics analyses by providing better sequence resolution, even in moderately complex regions (Borodinov et al., 2022).
Illumina's short-read sequencing excels in its accuracy, with Phred quality scores (Q-scores) commonly above Q30 along most of the read length (indicating an error rate of approximately <1%) (Jani et al., 2023; Thomas and Hahn, 2019). The PE strategy provides complementary reads at the two ends of the target DNA fragment, enabling error correction during PE base-calling and mitigating uncertainties. Additionally, the high sequencing depth of the platform guarantees significant read redundancy, which aids in detecting low-frequency variants and enables accurate allele identification. The confidence gained from such an analysis is largely due to the strength of the signal generated by the target gene, such as 12S rRNA, making it particularly sound for targeted sequencing applications (Woo et al., 2023). While read length has an inherent limit due to the chemistry of Illumina’s sequencing method, Illumina provides a great compromise of accuracy, throughput, and low-end cost that together positions its technology as highly suitable for use in 12S rRNA sequencing workflows (Woo et al., 2023).
Due to the acquisition of whole gene-spanning reads in a single sequence, long-read sequencing technologies like PacBio and ONT have significant advantages for analysing 12S rRNA genes (Cahyadi et al., 2020). Because of this extensive coverage, time-consuming assembly steps like merging overlapping reads, a necessity in Illumina workflows, are off the table. In particular, long-read sequencing is well suited for resolving structural variants or guanine-cytosine-rich (GC-rich) or repetitive regions of the 12S rRNA gene, as many short-read platforms often struggle to achieve adequate mapping in these regions (Wenger et al., 2019). In addition, long read sequencing can mitigate the biases in library preparation steps (for example, shearing and PCR amplification) that are commonly encountered in Illumina workflows (Bohmann et al., 2022). Historically, long-read technologies like ONT exhibit greater base-calling errors when compared to Illumina (Ma et al., 2019; Wick et al., 2023) though emerging technologies such as PacBio HiFi sequencing have brought this gap to a narrow. The long reads provided by PacBio HiFi now match the high-quality Phred scores of Illumina’s short reads, in addition to greater read length and accuracy.
The benefits of long-read sequencing in 12S rRNA sequencing are unequivocal for applications that require full-gene resolution (Zhou et al., 2023), such as species identification, genealogical analysis, or mitochondrial heteroplasmy detection. Long-read technologies afford full coverage of the target region and therefore contribute to the reduction of ambiguity and increase confidence in downstream analyses. Although short-read platforms are a cost-effective solution for high-throughput and targeted applications, long-read sequencing is indispensable for acquiring nuanced and complete sequence information for the 12S rRNA gene.
Both short- and long-read sequencing technologies have unique advantages for 12S rRNA sequencing. Illumina is the best option with respect to accuracy, cost efficiency, and high throughput capability, making it the best option for monogenic assays and identifying single variant genes. On the flip side, long-read technologies such as PacBio and ONT are needed for complete analyses requiring full-gene resolution and structural information and thus represent an evolution of meta-analysis capabilities within bioinformatics data analysis.
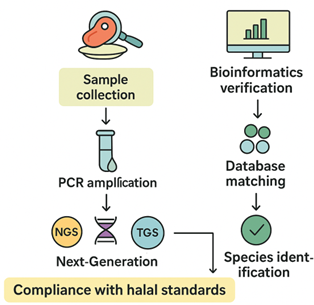
Figure 2: The streamlined workflow for molecular halal verification through mitochondrial 12S ribosomal RNA sequencing and bioinformatics analysis
NGS=Next Generation Sequencing; PCR=Polymerase Chain Reaction; TGS=Third Generation Sequencing
Figure 2 illustrates the streamlined workflow for molecular halal verification through mitochondrial 12S rRNA sequencing and bioinformatics analysis. The process begins with sample collection followed by PCR amplification of the 12S rRNA gene. The amplified DNA is then subjected to either NGS or TGS platforms to obtain high-resolution sequence data. The resulting sequences are processed through bioinformatics verification, where reads are matched against reference databases to achieve species identification. This molecular approach ensures accurate and reliable compliance with halal standards, enhancing transparency and traceability within the halal authentication process.
Streamlined sequencing for precision and reliability: advancing mitochondrial 12S rRNA analysis in industrial halal authentication
Next, because Illumina sequencing generates hundreds of short reads, the PE reads must be merged and undergo quality checks before being used in most downstream analytical tools, such as QIIME2 or Mothur (Meyer and Kircher, 2010). Thus, there is a step where overlapping read pairs are merged using special algorithms, followed by a quality-filtering step that removes low-quality bases and reads with performing errors. These operations provide a clean and reliable data source for the analyses that follow.
On the other hand, long-read sequencing systems, such as PacBio and ONT, directly sequence the amplified target region and generate long, continuous reads that cover the entire ROI (Wenger et al., 2019). Since these single reads span the entire targeted sequence, quality checking in long-read sequencing requires little verification. This streamlined pipeline reduces data preparation concerns, speeding up analytical operations and minimises the need for additional sequence processing.
The analysis of mitochondrial 12S rRNA sequencing consists of multiple interrelated steps and is facilitated by modern computational tools to ensure the reliability and reproducibility of results within a commercial halal authenticity testing pipeline (Yang et al., 2014). First, the pre-processing of data: Illumina sequenced data are trimmed and filtered to remove low-quality reads and adapter sequences. Bioinformatics pipelines typically apply tools such as Trimmomatic or Cutadapt at this point to clean the data and retain only high-quality reads for downstream analysis (Chen et al., 2018).
However, PacBio and ONT follow a similar systematic workflow, enabled by their long-read sequencing technologies. The ability to sequence the whole target region in one read, without the need for a read assembly, enhances the accuracy of variant detection (Uliano-Silva et al. 2023). Usually, after the quality check of Illumina, PacBio, and ONT platform data, the next step is to run software for downstream analysis such as QIIME2 or Mothur (Hernández and Guzmán, 2023). These tools help in taxonomic classification, sequence alignment, and variant detection. These are specific to Illumina's workflows and may range from different merging and quality-filtering of PE reads so as to get higher-quality short reads for further processing (Bolger et al., 2014). In contrast, both PacBio and ONT are capable of sequencing the entire 12S rRNA gene in a single read, which simplifies the pipeline and reduces the complexity of data interpretation (Roy et al., 2022). These platforms excel at resolving complex or repetitive regions, which can be quite challenging for short-read technologies.
All three technologies (Illumina, PacBio, ONT) have their own set of strengths. The continuous long fragments produced by PacBio and ONT improve the specificity and efficiency of 12S rRNA mitochondrial sequencing even though Illumina generates highly accurate short reads (El-Nabi et al., 2021). This means that the convergence of these technologies is especially well suited for the large-scale application of halal authenticity testing as it provides a comprehensive olive branch to large-scale sequencing capabilities.
Then reports are produced, making it very clear what the actual findings are, including what species have been detected, at what level of confidence, and where the halal profile appears to differ from the expected halal profile. They consist of Operational Taxonomic Units (OTUs) associated with each sample alongside descriptive data such as phylogenetic trees and relevant metadata (Venkatas and Adeleke, 2019). This increases the transparency and potential impact for stakeholders to utilise in making data-driven decisions from the analysis.
One of the first sequence data selected in a bioinformatics pipeline using the interspecies variability and intraspecies conservativeness of the 12S rRNA gene, a widely used mitochondrial marker provided strong resolution for distinguishing between species (Woo et al., 2023). This high-throughput approach, combining advanced sequencing technologies in a connected computational framework, offers assurances of precision and reproducibility.
Many pipelines are available to help run through the stages of the analysis. This has been carried out for quality control (Aksamentov et al., 2021), sequence alignment (Katoh et al., 2017), taxonomic classification (Menzel et al., 2016), Operational Taxonomic Unit (out) assignment (Wang et al., 2011) and final reporting (Taber, 2017). All pipelines were purpose-built to interface compatibly with the sequencing platform utilised (Wang et al., 2023) to enable data processing from initial sequencing reads to actionable results (Pillay et al., 2022). Other bioinformatics tools are also summarised in Table 6, which have been used for halal authentication analysis.
The table includes primers for multiple commonly consumed animals such as goat, chicken, cattle, sheep, horse, rat, dog, and pig. These species are frequently tested in halal verification to ensure the absence of non-halal ingredients or cross-contamination in food products. In addition, universal primers are included, which target conserved regions of the mitochondrial 12S rRNA gene and can amplify DNA across multiple vertebrate species.
Each primer set is associated with an expected amplicon size, which ranges from 155 bp (bovine) to 603 bp (rat), depending on the species and the genetic region targeted. Shorter amplicons are often preferred for processed food products, where DNA may be partially degraded. Longer amplicons may provide higher specificity in differentiating closely related species. For instance, the chicken primers produce amplicons of 227 bp and 596 bp, allowing flexibility depending on the sample type and quality.
The type of sample analysed using these primers can include raw meat, processed food, mixed meat products, or highly processed ingredients, all of which are relevant in halal authentication. The number of samples tested in each study may vary, but PCR amplification with these primers provides a rapid, sensitive, and species-specific method for detecting halal or non-halal components. For example, primers designed for pig DNA (non-halal) are critical for identifying contamination in food products labelled as halal.
References in Table 5 highlight the original sources where these primers were developed or validated, such as Matsunaga et al. (1999), Cahyadi et al. (2020), and Boyrusbianto et al. (2023). These studies demonstrated the reliability and reproducibility of these primers for molecular authentication. By integrating these primers into a PCR or sequencing workflow, halal verification can be performed accurately and efficiently, enabling both regulatory compliance and consumer confidence.
Overall, Table 5 illustrates the comprehensive set of primers available for PCR-based species identification, supporting the use of mtDNA markers such as 12S rRNA for halal authentication across a wide range of food products.
Optimising PCR amplification for high-quality sequencing across Illumina, PacBio, and ONT platforms
PCR amplification is a major process to enhance sequencing outputs for platforms such as Illumina, PacBio, and ONT. This step measures that the target ROI, like the 12S rRNA gene from genomic DNA (gDNA) input, is present in the proper quantity and quality, as both are essential for fetching high and reliable sequencing results (Kayama et al., 2021). Specific primers can be used to improve the selective amplification of target sequence, concentrating sequencing activity in the ROI and achieving high coverage, even for low-abundance regions (Ye et al., 2012).
The first steps of such a workflow typically involve the use of DNA extraction kits and protocols that protect DNA integrity (Dubois et al., 2022). From here, high-quality gDNA is used as an input for PCR amplification (Schloss et al., 2011). Next, primers are designed for the target ROI. For example, in the case of the 12S rRNA gene, primers are strategically chosen to bind conserved sequences flanking the target region, thereby clinching specificity (Woo et al., 2023). On the Illumina platform, primers are designed to amplify shorter DNA fragments (ranging from 250 to 300 bp) to ensure compatibility with the minimum read length required for Paired End (PE) sequencing (Kozich et al., 2013). On the other hand, large amplicon sequencing technologies (hundreds to a few kilobases), such as PacBio and ONT use primers designed to amplify the large amplicons, taking advantage of their long-read capabilities (Liu et al., 2020).
The cycle of denaturation, annealing, and extension is then repeated to achieve exponential amplification of the target genetic sequence (Nichols and Quince, 2016). The number of cycles is optimised based on the starting gDNA quantity and the desired amplicon length. Then, the amplified PCR products are purified to remove unincorporated primers, nucleotides, and other contaminants that might adversely affect sequencing. With long-read technologies, size selection is also applicable to ensure that the amplicons are of optimal size for sequencing. Depending on the platform, the next step of library preparation differs. For Illumina sequencing, the putative amplicons are ligated with platform-specific adapters, which ultimately form a sequencing library to be sequenced via PE or single-end (SE) strategies (Callahan et al., 2016). In a different approach, the PacBio and ONT libraries apply the ligation of adapters onto the amplified amplicons to prime them for long-read sequencing. Long-read platforms excel at resolving challenging areas, such as repeat elements and whole-gene analysis, including the 12S rRNA.
Once the sequencing libraries are ready, data generation begins. The second approach employed by Illumina is high-throughput, short-read sequencing, which delivers highly accurate data but often needs overlapping PE reads to cover the entire area of interest (Callahan et al., 2016). PacBio and ONT, on the other hand, use long-read sequencing methods that allow them to capture entire regions in a single pass, leading to easier assembly and more accurate mapping of challenging genomic regions. While gDNA serves the same fundamental role across all sequencing platforms, differences exist in the specific amplification and sequencing processes used (Woltyńska et al., 2023). PCR amplification enriches the signal of the desired sequence to achieve sufficient flow-cell coverage necessary for quality sequencing. Optimising workflows for respective platforms maximises sequencing potential, particularly in regions such as the 12S rRNA gene (Cahyadi et al., 2020). This includes Illumina short-read sequencing technology, which provides high throughput over small DNA fragments (Callahan et al., 2016). Meanwhile, long-read sequencing through PacBio and ONT offers better identification of structural variants and improved resolution of complex genomic segments (Ahsan et al., 2023).
To summarise, PCR amplification is an enabling step in sequencing workflows, enabling technologies, such as Illumina, PacBio, and ONT for a variety of applications. Non-Long Terminal Repeat (non-LTR) retrotransposons could certainly interfere with PCR amplification and reduce sequencing yields, though PCR remains essential for enriching target regions as well as ensuring a reliable sequence of interest. By implementing a bespoke amplification process, sequencing platforms have expanded their applications in species identification, phylogenetics, and mitochondrial genome studies (Schloss et al., 2011).
Short-read versus long-read sequencing: a comprehensive approach to 12S rRNA analysis for bioinformatics
It is widely known that Illumina sequencing technology boasts high throughput and outstanding accuracy, thus making it an optimal platform for producing short reads to cover target sequences, even when they do not exceed 500 bp (Kozich et al., 2013). Two-read analysis on mitochondrial 12S rRNA using Illumina’s PE sequencing format for 250 PE or 300 PE reads has been performed (Prakrongrak et al., 2023). However, the short-read property of Illumina sequencing makes it difficult to merge overlaps from PE reads simply, hence it typically needs to be preceded by tools like FLASH, PEAR, or BBMerge. These tools perform synchronisation and merging of the complementary reads to reconstruct the sequence of the entire target in question while ensuring that everything is covered correctly with minimal or no gaps or assembly variants. Such methods improve downstream bioinformatics analyses by providing better sequence resolution, even in moderately complex regions (Borodinov et al., 2022).
Illumina's short-read sequencing excels in its accuracy, with Phred quality scores (Q-scores) commonly above Q30 along most of the read length (indicating an error rate of approximately <1%) (Jani et al., 2023; Thomas and Hahn, 2019). The PE strategy provides complementary reads at the two ends of the target DNA fragment, enabling error correction during PE base-calling and mitigating uncertainties. Additionally, the high sequencing depth of the platform guarantees significant read redundancy, which aids in detecting low-frequency variants and enables accurate allele identification. The confidence gained from such an analysis is largely due to the strength of the signal generated by the target gene, such as 12S rRNA, making it particularly sound for targeted sequencing applications (Woo et al., 2023). While read length has an inherent limit due to the chemistry of Illumina’s sequencing method, Illumina provides a great compromise of accuracy, throughput, and low-end cost that together positions its technology as highly suitable for use in 12S rRNA sequencing workflows (Woo et al., 2023).
Due to the acquisition of whole gene-spanning reads in a single sequence, long-read sequencing technologies like PacBio and ONT have significant advantages for analysing 12S rRNA genes (Cahyadi et al., 2020). Because of this extensive coverage, time-consuming assembly steps like merging overlapping reads, a necessity in Illumina workflows, are off the table. In particular, long-read sequencing is well suited for resolving structural variants or guanine-cytosine-rich (GC-rich) or repetitive regions of the 12S rRNA gene, as many short-read platforms often struggle to achieve adequate mapping in these regions (Wenger et al., 2019). In addition, long read sequencing can mitigate the biases in library preparation steps (for example, shearing and PCR amplification) that are commonly encountered in Illumina workflows (Bohmann et al., 2022). Historically, long-read technologies like ONT exhibit greater base-calling errors when compared to Illumina (Ma et al., 2019; Wick et al., 2023) though emerging technologies such as PacBio HiFi sequencing have brought this gap to a narrow. The long reads provided by PacBio HiFi now match the high-quality Phred scores of Illumina’s short reads, in addition to greater read length and accuracy.
The benefits of long-read sequencing in 12S rRNA sequencing are unequivocal for applications that require full-gene resolution (Zhou et al., 2023), such as species identification, genealogical analysis, or mitochondrial heteroplasmy detection. Long-read technologies afford full coverage of the target region and therefore contribute to the reduction of ambiguity and increase confidence in downstream analyses. Although short-read platforms are a cost-effective solution for high-throughput and targeted applications, long-read sequencing is indispensable for acquiring nuanced and complete sequence information for the 12S rRNA gene.
Both short- and long-read sequencing technologies have unique advantages for 12S rRNA sequencing. Illumina is the best option with respect to accuracy, cost efficiency, and high throughput capability, making it the best option for monogenic assays and identifying single variant genes. On the flip side, long-read technologies such as PacBio and ONT are needed for complete analyses requiring full-gene resolution and structural information and thus represent an evolution of meta-analysis capabilities within bioinformatics data analysis.

Figure 2: The streamlined workflow for molecular halal verification through mitochondrial 12S ribosomal RNA sequencing and bioinformatics analysis
NGS=Next Generation Sequencing; PCR=Polymerase Chain Reaction; TGS=Third Generation Sequencing
Figure 2 illustrates the streamlined workflow for molecular halal verification through mitochondrial 12S rRNA sequencing and bioinformatics analysis. The process begins with sample collection followed by PCR amplification of the 12S rRNA gene. The amplified DNA is then subjected to either NGS or TGS platforms to obtain high-resolution sequence data. The resulting sequences are processed through bioinformatics verification, where reads are matched against reference databases to achieve species identification. This molecular approach ensures accurate and reliable compliance with halal standards, enhancing transparency and traceability within the halal authentication process.
Streamlined sequencing for precision and reliability: advancing mitochondrial 12S rRNA analysis in industrial halal authentication
Next, because Illumina sequencing generates hundreds of short reads, the PE reads must be merged and undergo quality checks before being used in most downstream analytical tools, such as QIIME2 or Mothur (Meyer and Kircher, 2010). Thus, there is a step where overlapping read pairs are merged using special algorithms, followed by a quality-filtering step that removes low-quality bases and reads with performing errors. These operations provide a clean and reliable data source for the analyses that follow.
On the other hand, long-read sequencing systems, such as PacBio and ONT, directly sequence the amplified target region and generate long, continuous reads that cover the entire ROI (Wenger et al., 2019). Since these single reads span the entire targeted sequence, quality checking in long-read sequencing requires little verification. This streamlined pipeline reduces data preparation concerns, speeding up analytical operations and minimises the need for additional sequence processing.
The analysis of mitochondrial 12S rRNA sequencing consists of multiple interrelated steps and is facilitated by modern computational tools to ensure the reliability and reproducibility of results within a commercial halal authenticity testing pipeline (Yang et al., 2014). First, the pre-processing of data: Illumina sequenced data are trimmed and filtered to remove low-quality reads and adapter sequences. Bioinformatics pipelines typically apply tools such as Trimmomatic or Cutadapt at this point to clean the data and retain only high-quality reads for downstream analysis (Chen et al., 2018).
However, PacBio and ONT follow a similar systematic workflow, enabled by their long-read sequencing technologies. The ability to sequence the whole target region in one read, without the need for a read assembly, enhances the accuracy of variant detection (Uliano-Silva et al. 2023). Usually, after the quality check of Illumina, PacBio, and ONT platform data, the next step is to run software for downstream analysis such as QIIME2 or Mothur (Hernández and Guzmán, 2023). These tools help in taxonomic classification, sequence alignment, and variant detection. These are specific to Illumina's workflows and may range from different merging and quality-filtering of PE reads so as to get higher-quality short reads for further processing (Bolger et al., 2014). In contrast, both PacBio and ONT are capable of sequencing the entire 12S rRNA gene in a single read, which simplifies the pipeline and reduces the complexity of data interpretation (Roy et al., 2022). These platforms excel at resolving complex or repetitive regions, which can be quite challenging for short-read technologies.
All three technologies (Illumina, PacBio, ONT) have their own set of strengths. The continuous long fragments produced by PacBio and ONT improve the specificity and efficiency of 12S rRNA mitochondrial sequencing even though Illumina generates highly accurate short reads (El-Nabi et al., 2021). This means that the convergence of these technologies is especially well suited for the large-scale application of halal authenticity testing as it provides a comprehensive olive branch to large-scale sequencing capabilities.
Then reports are produced, making it very clear what the actual findings are, including what species have been detected, at what level of confidence, and where the halal profile appears to differ from the expected halal profile. They consist of Operational Taxonomic Units (OTUs) associated with each sample alongside descriptive data such as phylogenetic trees and relevant metadata (Venkatas and Adeleke, 2019). This increases the transparency and potential impact for stakeholders to utilise in making data-driven decisions from the analysis.
One of the first sequence data selected in a bioinformatics pipeline using the interspecies variability and intraspecies conservativeness of the 12S rRNA gene, a widely used mitochondrial marker provided strong resolution for distinguishing between species (Woo et al., 2023). This high-throughput approach, combining advanced sequencing technologies in a connected computational framework, offers assurances of precision and reproducibility.
Many pipelines are available to help run through the stages of the analysis. This has been carried out for quality control (Aksamentov et al., 2021), sequence alignment (Katoh et al., 2017), taxonomic classification (Menzel et al., 2016), Operational Taxonomic Unit (out) assignment (Wang et al., 2011) and final reporting (Taber, 2017). All pipelines were purpose-built to interface compatibly with the sequencing platform utilised (Wang et al., 2023) to enable data processing from initial sequencing reads to actionable results (Pillay et al., 2022). Other bioinformatics tools are also summarised in Table 6, which have been used for halal authentication analysis.
Table 6: Bioinformatics tools for metagenomics analysis in halal authentication
| Tools | Details | References |
| CLAssifier based on Reduced K-mers (CLARK) | A fast and accurate classifier for metagenomics sequences using k-mer reduction for taxonomic identification. | (Ounit et al., 2015) |
| Kraken 2 | An ultrafast, accurate metagenomics classifier that uses exact k-mer matches for taxonomic assignment. | (Wood et al., 2019) |
| Metagenomic Phylogenetic Analysis (MetaPhlAn) | Profiles microbial community composition using marker genes, providing taxonomic resolution. | (Segata et al., 2012) |
| Bioinformatics for Microbial Sequences (BioMaS) | BioMaS pipeline for taxonomic profiling of microbiomes in clinical and environmental studies. | (Fosso et al., 2015) |
| All-Food-Seq (AFS) | A pipeline for detecting and typing pathogens in food samples, supporting food-borne pathogen surveillance. | (Ripp et al., 2014) |
| MGnify | A resource provided by the European Molecular Biology Laboratory's European Bioinformatics Institute (EMBL-EBI) for assembly, annotation, and analysis of metagenomics datasets. | (Mitchell et al., 2020) |
Bioinformatics is a highly flexible and adaptable domain, depending on the sequencing of the minutest details from the input sequence all the way to the platform used for sequencing. Platform selection will heavily shape the analysis workflow, as the analytical demands are disparate for short-read platforms such as Illumina versus long-read options like PacBio or ONT (Callahan et al., 2016). But no matter which vessel is chosen, or the method of presentation, careful attention must be given to the quality and quantity of the output readings. Having high-quality data and coverage is essential for accurate analysis because low-quality reads and sequencing depth can lead to incomplete or untrustworthy results (Shirazi et al., 2021). Hence, careful implementations of quality assurance and optimisation of bioinformatics processes are essential for providing credible and reproducible results.
Food and safety in halal context
The introduction of innovative molecular approaches for halal authentication highlights their importance in ensuring food safety. This approach provides a holistic method to make sure that halal foods are wholesome and safe, at the expense of religious dogma and firm science. At the same time, the contents of food products should be validated to determine whether they are authentic and whether they originate from the claimed sources (Daniel et al. 2022). The present study emphasises mitochondrial 12S rRNA sequencing (El-Nabi et al., 2021) as an approach that strikes at the heart of safety concerns associated with both intentional or unintentional contamination of halal foods with haram components.
Visual or manual inspection methodologies are incapable of guaranteeing food safety, and as such, molecular methodologies are crucially necessary in food processing. The risk of cross-contamination or false labelling is eradicated, by detecting even the slightest trace of a non-halal ingredient. For consumers, this is even more significant, especially those with dietary restrictions or allergies, which make it critical to avoid harmful substances (Hidayat et al., 2023). Food is manufactured and spread around the world, and this globalised nature creates huge risks of contaminants and safety breaches. Ensuring that ingredients, manufacturing processes, and handling conform to both safety standards and religious beliefs is particularly challenging in the realm of halal food safety. With the combinations of NGS and bioinformatics methods, a complete identification of contamination of the whole supply chain is presented (Fu et al., 2012).
The techniques described here are not only used to ascertain the truth of a species but also to characterise microbiological contaminants, which guarantee the good of food products from horrible infections (Delahoy et al., 2023). This dual molecular technology protects against foodborne infections, which are a serious global public health concern, while also complying with halal certification (Krisna and Yusuf, 2023). Hence, cleanliness and hygiene are key elements of halal food safety, built on the foundation of Tayyib, which emphasises purity and wholesomeness (Aksin and Aini, 2023).
Best practices to ensure halal traceability
This review represents significant progress in the halal supply chain, utilising technology, accountability, and transparency. Indeed, traceability systems are essential for maintaining food safety, enabling stakeholders to trace ingredients from farm to fork and guarantee that all relevant parties in the supply chain comply with regulation (Bosona and Gebresenbet, 2023). Technology provides an additional level of transparency that makes it possible to detect and respond quickly to contamination events and safety violations.
This then results in the integration of recent scientific findings with accepted religious guidelines and makes meaningful contributions toward the discussion within the halal paradigm. Their suggested approaches pave the way for ethical and technological inclusion in the halal space while successfully addressing the intricacies of modern supply chains (Cobbe et al., 2023). Fighting these battles will require innovation and embracing creative solutions in doing so is critical. This will serve as a cornerstone for preserving the integrity of halal-certified products, as it guarantees their safety and reliability as global consumption continues to rise (Oktavia et al., 2019).
Future perspectives and applications of halal products
The above-mentioned approach demonstrates how full utilisation of integrated state-of-the-art technologies such as multi-omics approaches is essential to enhance compliance, quality assurance, and transparency standards in the halal industry (Pang et al., 2022). This vision serves as a reflection of the growing worldwide focus on halal products, while also highlighting the need for technology to meet consumer demands and industry standards (Ezeaku et al., 2021).
It is a compelling technical approach in that it links leading-edge scientific innovations to practical solutions. For instance, the advent of portable sequencing devices as well as in-field, automated data analysis tools provides a compelling pathway for efficiency gains and lower barriers to authentication validation to stakeholders (Hayes et al., 2021). In addition, the request for common sequencing technologies and standardised analysis pipelines demonstrates a deeper understanding of the challenges to harmonisation of methods and maintaining comparability across many disparate regions and laboratories (Algan, 2023).
A particularly compelling link exists between halal authenticity and food safety. Based on the technicalities of 12S rRNA sequencing, which resonate with the general theme of well-being in a consumer era, the success rests on the identification of commonalities around hygiene to prevent contamination and assurance of computation quality (Kumari et al., 2019). The complementarity of both aspects signifies the need for a joint application of halal compliance and food safety, strengthening the underlying rationale of using modern technologies for these aims.
The ensuing discussion should include the practical implementation of these metagenomic and DNA approaches in halal testing pipelines (Cantalapiedra et al. 2021). Similarly, accounting for complicating factors, such as the cost and plausibility of incorporating technologies like portable sequencing devices would provide a clearer picture (Hayes et al. 2021). Another way to improve reliability would be by directly correlating ethics with issues arising from data privacy in consumer platforms technology (Gallardo et al., 2023; Sanjaya and Arabella, 2023).
Moreover, including practical examples and/or case studies from other industries with similar traceability needs (e.g., kosher certification and broader food safety needs) would further help readers understand the practicability of the proposals (Nicolae et al., 2017). Addressing specific omissions in existing halal certification across regions may also provide a strong justification for the proposed standardisation efforts (Yakin, 2021). Lastly, capturing the interest of readers by focusing on the sustainability aspect of these technologies for instance, their economic and environmental advantages, such as the reduction of waste or the optimisation of resources utilised in halal production, would further strengthen the discussion (Allam et al., 2022; Jiang, 2023). One of its significant strengths is its consumer-driven, privacy-preserving solutions (e.g., platforms that allow consumers to directly access the result of the authenticity test) (Muralidharan et al., 2023). By emphasising transparency, these solutions contribute to enhancing consumer trust, an important component in the global halal space (Talib and Zulfakar, 2023). However, some thoughts on how these platforms could be created, implemented, and maintained (especially in low-resource settings), would provide a practical dimension to this claim (Mathew, 2023).
Conclusion
The industrial-scale DNA sequencing technologies discussed in this article support halal authentication, thereby enabling stakeholders and consumers to achieve greater transparency, compliance, and trust. To address concerns regarding authenticity and traceability issues in halal, this study utilised modern sequencing techniques as a credible means for validating halal product integrity. It also serves to reinforce the moral aspects within the supply chain as well as to encourage the growth and long-term viability of the halal industry. Consequently, the industry is increasingly adopting advanced methods such as DNA sequencing to provide consumers with verified and up-to-date verification of a product's halal status. This is important in combating fraud, mislabelling, and contamination, thereby supporting the religious and cultural preferences of consumers.
In addition, the global acceleration of commerce and communication has strengthened the need for halal standardisation of halal regulations on an international scale. Industrial-scale sequencing technology supports this by offering companies a simple solution to cross-navigate the heterogeneous regulatory landscape in the jurisdictions through platforms designed to standardise and unify the halal certification process. The broader long-term benefits of this approach certainly outweigh its cost. However, the technical sophistication does come with its own costs. Nevertheless, the technology complements consumer trust, reinforces brand equity, and aligns with the moral and religious beliefs of consumers, thus nurturing the sustainable growth of the halal industry. Moreover, sequencing technology provides transparency that caters to the increasing demand for information and ethical practices in today’s food system.
Author contributions
J.J. and M.A.S. designed the study; J.J., M.A.S, C.L.S.L. and S.M. collected the data J.J., M.A.S, C.L.S.L. and S.M. analyzed the data and wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgements
The authors are grateful to the team members for their contributions of ideas, knowledge, and financial support, especially Constance and Symeon from the Department of Anesthesiology and Intensive Care, Faculty of Medicine and Health Sciences, Kota Kinabalu, Sabah, Malaysia, and from the School of Business, Swinburne University of Technology, Kuching, Sarawak.
Conflicts of interest
The authors declare that there is no conflict of interest.
Funding
This research received no specific grant from any funding agency in the public, commercial, or non-profit sectors.
Ethical consideration
Not applicable.
References
Absmeier E., Chandrasekaran V., O’Reilly F.J., Stowell J.a.W., Rappsilber J., Passmore L.A. (2023). Specific recognition and ubiquitination of translating ribosomes by mammalian CCR4–NOT. Nature Structural and Molecular Biology. 30: 1314–1322. [DOI: 10.1038/s41594-023-01075-8]
Ahsan M.U., Liu Q., Perdomo J.E., Fang L., Wang K. (2023). A survey of algorithms for the detection of genomic structural variants from long-read sequencing data. Nature Methods. 20: 1143–1158. [DOI: 10.1038/s41592-023-01932-w]
Aini S.R., Rohman A., Mulyanto M., Erwanto Y., Ansar A., Handayani B.R., Irnawati I. (2023). The metabolomics approach used for halal authentication analysis of food and pharmaceutical products: a review. Food Research. 7: 180–187.
Aksamentov I., Roemer C., Hodcroft E.B., Neher R.A. (2021). Nextclade: clade assignment, mutation calling and quality control for viral genomes. Journal of Open Source Software. 6: 3773. [DOI: 10.21105/joss.03773]
Aksin N., Aini F.N.Q. (2023). Halal Ṭayyib syaria’ in understanding the needs of consumer protection. Jurnal Meta-Yuridis. 6: 1–13. [DOI: 10.26877/m-y.v6i2.15560]
Algan F.M. (2023). Global standardization for a sustainable future. Current Research in Social Sciences. 9: 30-40. [DOI: 10.30613/ curesosc.1173926]
Allam Z., Bibri S.E., Chabaud D., Moreno C. (2022). The theoretical, practical, and technological foundations of the 15-minute city model: proximity and its environmental, social, and economic benefits for sustainability. Energies. 15: 6042. [DOI: 10.3390/ en15166042]
Amarasinghe S.L., Su S., Dong X., Zappia L., Ritchie M.E., Gouil Q. (2020). Opportunities and challenges in long-read sequencing data analysis. Genome Biology. 21: 30. [DOI: 10.1186/s13059-020-1935-5]
Amer M. (2024). Halal standards’ implementation in Palestinian food sector: its drivers and impact on performance. Arab Gulf Journal of Scientific Research. 42: 2–29. [DOI: 10.1108/agjsr-09-2022-0168]
Aziz N., Bakry N., Habibi Mz M., Siddiq Armia M. (2023). The paradigm of modern food products and its relevance with the concept of food in the Quran. Heliyon. 9: e21358. [DOI: 10.1016/j. heliyon.2023.e21358]
Bannor R.K., Arthur K.E., Oppong D., Oppong-Kyeremeh H. (2023). A comprehensive systematic review and bibliometric analysis of food fraud from a global perspective. Journal of Agriculture and Food Research. 14: 100686. [DOI: 10.1016/j.jafr. 2023.100686]
Bianconi I., Aschbacher R., Pagani E. (2023). Current uses and future perspectives of genomic technologies in clinical microbiology. Antibiotics. 12: 1580. [DOI: 10.3390/antibiotics12111580]
Bohmann K., Elbrecht V., Carøe C., Bista I., Leese F., Bunce M., Yu D.W., Seymour M., Dumbrell A.J., Creer S. (2022). Strategies for sample labelling and library preparation in DNA metabarcoding studies. Molecular Ecology Resources. 22: 1231–1246. [DOI: 10.1111/1755-0998.13512]
Bolger A.M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30: 2114–2120. [DOI: 10.1093/bioinformatics/btu170]
Borodinov A., Manoilov V., Zarutsky I., Petrov A., Kurochkin V., Saraev A. (2022). Machinelearning in base-calling for next-generation sequencing methods. Informatics and Automation. 21: 572–603. [DOI: 10.15622/ia.21.3.5]
Bosona T., Gebresenbet G. (2023). The role of blockchain technology in promoting traceability systems in agri-food production and supply chains. Sensors. 23: 5342. [DOI: 10.3390/s23115342]
Boyrusbianto F.T., Utama D.T., Khikmawati I., Fadhila A., Volkandari S.D., Abdurrahman Z.H., Pramono A., Cahyadi M. (2023). A simplex and multiplex PCR assay for simultaneous detection of beef, pork, and chicken meat in sausages based on mitochondrial DNA Cytochrome oxidase sub-unit I. Food Research. 7: 188–193. [DOI: 10.26656/fr.2017.7(3).084]
Cahyadi M., Wibowo T., Pramono A., Abdurrahman Z.H. (2020). A novel multiplex-PCR assay to detect three non-halal meats contained in meatballs using mitochondrial 12S rRNA gene. Food Science of Animal Resources. 40: 628–635. [DOI: 10.5851/kosfa.2020.e40]
Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nature Methods. 13: 581–583. [DOI: 10.1038/ nmeth.3869]
Cantalapiedra C.P., Hernández-Plaza A., Letunic I., Bork P., Huerta-Cepas J. (2021). eggNOG-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Molecular Biology and Evolution. 38: 5825-5829. [DOI: 10.1093/molbev/msab293]
Cao M.D., Ganesamoorthy D., Cooper M.A., Coin L.J.M. (2015). Realtime analysis and visualization of MinION sequencing data with npReader. Bioinformatics. 32: 764–766. [DOI: 10.1093/bioinformatics/btv658]
Chan A.H.E., Saralamba N., Saralamba S., Ruangsittichai J., Thaenkham U. (2022). The potential use of mitochondrial ribosomal genes (12S and 16S) in DNA barcoding and phylogenetic analysis of trematodes. BMC Genomics. 23: 104. [DOI: 10.1186/s12864-022-08302-4]
Chang J.J.M., Ip Y.C.A., Bauman A.G., Huang D. (2020). MINION-in-ARMS: nanopore sequencing to expedite barcoding of specimen-rich macrofaunal samples from autonomous reef monitoring structures. Frontiers in Marine Science. 7: 448. [DOI: 10.3389/fmars.2020. 00448]
Chen C., Wu Y., Li J., Wang X., Zeng Z., Xu J., Liu Y., Feng J., Chen H., He Y., Xia R. (2023). TBtools-II: a "one for all, all for one" bioinformatics platform for biological big-data mining. Molecular Plant. 16: 1733-1742. [DOI: 10.1016/j.molp.2023.09.010]
Chen S., Zhou Y., Chen Y., Gu J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34: i884–i890. [DOI: 10.1093/bioinformatics/bty560]
Cobbe J., Veale M., Singh J. (2023). Understanding accountability in algorithmic supply chains. Proceedings of the 2023 ACM Conference on Fairness, Accountability, and Transparency. 1186–1197. [DOI: 10.1145/3593013.3594073]
Daniel A.I., Fadaka A.O., Gokul A., Bakare O.O., Aina O., Fisher S., Burt A.F., Mavumengwana V., Keyster M., Klein A. (2022). Biofertilizer: the future of food security and food safety. Microorganisms. 10: 1220. [DOI: 10.3390/microorganisms10061220]
De Coster W., Weissensteiner M.H., Sedlazeck F.J. (2021). Towards population-scale long-read sequencing. Nature Reviews Genetics. 22: 572–587. [DOI: 10.1038/s41576-021-00367-3]
Delahoy M.J., Shah H.J., Weller D.L., Ray L.C., Smith K., McGuire S., Trevejo R.T., Walter E.S., Wymore K., Rissman T., McMillian M., Lathrop S., et al. (2023). Preliminary incidence and trends of infections caused by pathogens transmitted commonly through food — foodborne diseases active surveillance network, 10 U.S. sites, 2022. Morbidity and Mortality Weekly Report. 72: 701–706. [DOI: 10.15585/ mmwr.mm7226a1]
Dubois B., Debode F., Hautier L., Hulin J., Martin G.S., Delvaux A., Janssen E., Mingeot D. (2022). A detailed workflow to develop QIIME2-formatted reference databases for taxonomic analysis of DNA metabarcoding data. BMC Genomic Data. 23: 53. [DOI: 10.1186/s12863-022-01045-w]
Dunham J.P., Friesen M.L. (2013). A cost-effective method for high-throughput construction of Illumina sequencing libraries. Cold Spring Harbor Protocols. 2013: 820-834. [DOI: 10.1101/pdb.prot074187]
El-Nabi S.E.H., Hussein D., Khallafa A.G. (2021). Molecular detection of food fraud targeting mitochondrial 12S rRNA gene sequencing. Journal of Bioscience and Applied Research. 7: 17–22. [DOI: 10.21608/jbaar.2021.167091]
Ezeaku H.C., Asongu S.A., Nnanna J. (2021). Volatility of international commodity prices in times of COVID-19: effects of oil supply and global demand shocks. The Extractive Industries and Society. 8: 257-270. [DOI: 10.1016/j.exis.2020.12.013]
Ferreira T., Rodriguez S. (2024). Mitochondrial DNA: inherent complexities relevant to genetic analyses. Genes. 15: 617. [DOI: 10.3390/ genes15050617]
Fosso B., Santamaria M., Marzano M., Alonso-Alemany D., Valiente G., Donvito G., Monaco A., Notarangelo P., Pesole G. (2015). BioMaS: a modular pipeline for bioinformatic analysis of metagenomic amplicons. BMC Bioinformatics. 16: 203. [DOI: 10.1186/s12859-015-0595-z]
Fu L., Niu B., Zhu Z., Wu S., Li W. (2012). CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 28: 3150–3152. [DOI: 10.1093/bioinformatics/bts565]
Galal-Khallaf A. (2021). Multiplex PCR and 12S rRNA gene sequencing for detection of meat adulteration: a case study in the Egyptian markets. Gene. 764: 145062. [DOI: 10.1016/j.gene.2020.145062]
Gallardo A., Choy C., Juneja J., Bozkir E., Cobb C., Bauer L., Cranor L. (2023). Speculative privacy concerns about AR glasses data collection. Proceedings on Privacy Enhancing Technologies. 2023: 416-435. [DOI: 10.56553/ popets-2023-0117]
Garinet S., Laurent-Puig P., Blons H., Oudart J. (2018). Current and future molecular testing in NSCLC: what can we expect from new sequencing technologies? Journal of Clinical Medicine. 7: 144. [DOI: 10.3390/jcm7060144]
Gupta N., Verma V. (2019). Next-generation sequencing and its application: empowering in public health beyond reality. In: Arora P. (Editor). Microbial technology for the welfare of society. Springer, Singapore. pp: 313–341. [DOI: 10.1007/978-981-13-8844-6_15]
Hassan N., Ahmad T., Zain N.M., Ashaari A. (2020). A novel chemometrics method for halal authentication of gelatin in food products. SainsMalaysiana. 49: 2083–2089. [DOI: 10.17576/jsm-2020-4907-08]
Hayes B., Nguyen L.T., Forutan M., Engle B.N., Lamb H.J., Copley J.P., Randhawa I.A.S., Ross E.M. (2021). An epigenetic aging clock for cattle using portable sequencing technology. Frontiers in Genetics. 18: 760450. [DOI: 10.3389/fgene. 2021.760450]
Haynes E., Jiménez E., Pardo M.Á., Helyar S.J. (2019). The future of NGS (Next Generation Sequencing) analysis in testing food authenticity. Food Control. 101: 134–143. [DOI: 10.1016/j.foodcont. 2019.02. 010]
Hernández V.P., Guzmán M.H. (2023). Microorganisms identification in environmental samples: bioinformatic analysis of 16S rRNA gene with QIIME2. Ciencia Latina Revista Científica Multidisciplinar. 7: 8074–8102. [DOI: 10.37811/cl_rcm.v7i5.8382]
Hidayat H.H., Wijayanti N., Situmorang S.C.D.U.B. (2023). Materials clustering of Soto Sokaraja as an effort to accelerate halal certification. IOP Conference Series: Earth and Environmental Science. 1177: 012036. [DOI: 10.1088/1755-1315/1177/ 1/012036]
Hook P.W., Timp W. (2023). Beyond assembly: the increasing flexibility of single-molecule sequencing technology. Nature Reviews Genetics. 24: 627–641. [DOI: 10.1038/s41576-023-00600-1]
Jani N.J., Chin N.K.L., Mustapha N.Z.A. (2023). Whole genome sequence analysis of a Mycobacterium iranicum, a newly identified non-tuberculosis mycobacteria, strain SBH312 isolated in Sabah, Malaysia. Borneo Journal of Medical Sciences. 17: 52–56. [DOI: 10.51200/bjms.v17i1.4291]
Jaswir I., Mirghani M.E.S., Salleh H.M., Ramli N., Octavianti F., Hendri R. (2016). An overview of the current analytical methods for halal testing. Current issues in halal food analysis. Springer, Singapore. pp: 291–300. [DOI: 10.1007/978-981-10-1452-9_27]
Jiang L. (2023). Environmental benefits of green buildings with BIM technology. Ecological Chemistry and Engineering. 30: 191-199. [DOI: 10.1515/eces-2023-0003]
Jovic D., Liang X., Zeng H., Lin L., Xu F., Luo Y. (2022). Single‐cell RNA sequencing technologies and applications: a brief overview. Clinical and Translational Medicine. 12: 21-35. [DOI: 10.1002/ ctm2.983]
Karst S.M., Ziels R.M., Kirkegaard R., Sørensen E.A., McDonald D., Zhu Q., Knight R., Albertsen M. (2021). High-accuracy long-read amplicon sequences using unique molecular identifiers with Nanopore or PacBio sequencing. Nature Methods. 18: 165-169. [DOI: 10.1038/s41592-020-01041-y]
Katoh K., Rozewicki J., Yamada K.D. (2017). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics. 20: 1160–1166. [DOI: 10.1093/bib/bbx108]
Kaveh S., Hashemi S.M.B., Abedi E., Amiri M.J., Conte F.L. (2023). Bio-preservation of meat and fermented meat products by lactic acid bacteria strains and their antibacterial metabolites. Sustainability. 15: 10154. [DOI: 10.3390/su151310154]
Kayama K., Kanno M., Chisaki N., Tanaka M., Yao R., Hanazono K., Camer G.A., Endoh D. (2021). Prediction of PCR amplification from primer and template sequences using recurrent neural network. Scientific Reports. 11: 7493. [DOI: 10.1038/s41598-021-86357-1]
King J., Harder T., Beer M., Pohlmann A. (2020). Rapid multiplex MinION nanopore sequencing workflow for Influenza A viruses. BMC Infectious Diseases. 20: 648. [DOI: 10.1186/s12879-020-05367-y]
Kozich J.J., Westcott S.L., Baxter N.T., Highlander S.K., Schloss P.D. (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MISEQ Illumina sequencing platform. Applied and Environmental Microbiology. 79: 5112–5120. [DOI: 10.1128/aem. 01043-13]
Krisna R., Yusuf M. (2023). Halal ecosystem improvement study reviewed of halal product regulations. International Journal of Research and Review. 10: 339–353. [DOI: 10.52403/ijrr.20230243]
Kumari P., Dong K., Eo K., Lee W., Kimura J., Yamamoto N. (2019). DNA metabarcoding-based diet survey for the Eurasian otter (Lutralutra): Development of a Eurasian otter-specific blocking oligonucleotide for 12S rRNA gene sequencing for vertebrates. PLOS ONE. 14: e0226253. [DOI: 10.1371/journal.pone.0222966]
Liu H., Nie J., Liu Y., Wadood S.A., Rogers K.M., Yuan Y., Gan R. (2023). A review of recent compound-specific isotope analysis studies applied to food authentication. Food Chemistry. 415: 135791. [DOI: 10.1016/j.foodchem.2023.135791]
Liu J., Seetharam A.S., Chougule K., Ou S., Swentowsky K.W., Gent J.I., Llaca V., Woodhouse M.R., Manchanda N., Presting G.G., Kudrna D.A., Alabady M., Hirsch C.N., Fengler K.A., Ware D., Michael T.P., Hufford M.B., Dawe R.K. (2020). Gapless assembly of maize chromosomes using long-read technologies. Genome Biology. 21: 121. [DOI: 10.1186/s13059-020-02029-9]
Liu X., Liu Z., Cheng Y., Wu H., Shen W., Liu Y., Feng Q., Yang J. (2021). Application of next-generation sequencing technology based on single gene locus in species identification of mixed meat products. Journal of Food Quality. 2021: 4512536. [DOI: 10. 1155/2021/4512536]
Lu H., Giordano F., Ning Z. (2016). Oxford NanoporeMinION sequencing and genome assembly. Genomics, Proteomics and Bioinformatics. 14: 265–279. [DOI: 10.1016/j.gpb.2016.05.004]
Lubis H., Mohd-Naim N.F., Alizul N.N., Ahmed M.U. (2016). From market to food plate: current trusted technology and innovations in halal food analysis. Trends in Food Science and Technology. 58: 55–68. [DOI: 10.1016/j.tifs.2016.10.024]
Lücking R., Aime M.C., Robbertse B., Miller A., Ariyawansa H., Aoki T., Papp V., Robert V., Schoch C. (2020). Unambiguous identification of fungi: where do we stand and how accurate and precise is fungal DNA barcoding? IMA Fungus. 11: 14. [DOI: 10.1186/s43008-020-00033-9]
Luo J., Yan D., Zhang D., Han Y., Dong X., Yang Y., Deng K., Xiao X. (2011). Application of 12S rRNA gene for the identification of animal-derived drugs. Journal of Pharmacy and Pharmaceutical Sciences. 14: 358–367. [DOI: 10.18433/j3n017]
Ma X., Shao Y., Tian L., Flasch D.A., Mulder H.L., Edmonson M.N., Liu Y., Chen X., Newman S., Nakitandwe J., Li Y., Li B., Shen S., Wang Z., Shurtleff S., Robison L.L., Levy S., Easton J., Zhang J. (2019). Analysis of error profiles in deep next-generation sequencing data. Genome Biology. 20: 50. [DOI: 10.1186/s13059-019-1659-6]
Malaysia’s Halal Food Opportunities (MIDA). (2022). Malaysia’s halal food opportunities - carving out the lion’s share in a USD3 trillion global market. URL: https://www.mida.gov.my/malaysias-halal-food-opportunitiescarving-out-the-lions-share-in-a-usd3-trillion-global-market/. Accessed on 5 January 2025.
Mathew J.L. (2023). Efficiency, economy, and excellence: experimental exploration of evidence-based guideline development in resource-constrained settings. Indian Journal of Pediatrics. 90: 700-707. [DOI: 10.1007/s12098-023-03785-1]
Matsunaga T., Chikuni K., Tanabe R., Muroya S., Shibata K., Yamada J., Shinmura Y. (1999). A quick and simple method for the identification of meat species and meat products by PCR assay. Meat Science. 51: 143–148. [DOI: 10.1016/s0309-1740(98)00112-0]
Menzel P., Ng K.L., Krogh A. (2016). Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nature Communications. 7: 11257. [DOI: 10.1038/ncomms11257]
Meyer M., Kircher M. (2010). Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harbor Protocols. 2010: pdb.prot5448. [DOI: 10.1101/pdb.prot5448]
Mitchell A.L., Almeida A., Beracochea M., Boland M., Burgin J., Cochrane G., Crusoe M.R., Kale V., Potter S.C., Richardson L.J., Sakharova E., Scheremetjew M., et al. (2020). MGnify: the microbiome analysis resource in 2020. Nucleic Acids Research. 48: D570–D578. [DOI: 10.1093/nar/gkz1035]
Moorthie S., Mattocks C., Wright C.F. (2011). Review of massively parallel DNA sequencing technologies. The HUGO Journal. 5: 1–12. [DOI: 10.1007/s11568-011-9156-3]
Montiel-Sosa J.F., Ruiz-Pesini E., Montoya J., Roncalés P., López-Pérez M.J., Pérez-Martos A. (2000). Direct and highly species-specific detection of pork meat and fat in meat products by PCR amplification of mitochondrial DNA. Journal of Agricultural and Food Chemistry. 48: 2829–2832. [DOI: 10.1021/jf9907438]
Monzel A.S., Enríquez J.A., Picard M. (2023). Multifaceted mitochondria: moving mitochondrial science beyond function and dysfunction. Nature Metabolism. 5: 546–562. [DOI: 10.1038/s42255-023-00783-1]
Muflihah, Hardianto A., Kusumaningtyas P., Prabowo S., Hartati Y.W. (2023). DNA-based detection of pork content in food. Heliyon. 9: e14418. [DOI: 10.1016/j.heliyon.2023.e14418]
Muralidharan S., Yoo B., Ko H. (2023). Decentralized ME-centric framework—a futuristic architecture for consumer IoT. IEEE Consumer Electronics Magazine. 12: 39-50. [DOI: 10.1109/ MCE. 2023.3152809]
Murugaiah C., Noor Z.M., Mastakim M., Bilung L.M., Selamat J., Radu S. (2009). Meat species identification and halal authentication analysis using mitochondrial DNA. Meat Science. 83: 57–61. [DOI: 10.1016/j.meatsci.2009.03.015]
Nasir S., Zulfakar M.H., Rahmat A. (2021). Determinants of preferred export logistics gateway in Malaysia halal product industry. International Journal of Academic Research in Accounting, Finance and Management Sciences. 11: 60-69. [DOI: 10.6007/IJARAFMS/v11-i4/11823]
Neequaye G., Agyekum K., Botchway E. (2017). Market analysis of halal food consumption. International Journal of Food Science. 52: 112-124. [DOI: 10.1002/ijfs.2017.112]
Ng P.C., Ruslan N.A.S.A., Chin L.X., Ahmad M., Hanifah S.A., Abdullah Z., Khor S.M. (2021). Recent advances in halal food authentication: challenges and strategies. Journal of Food Science. 87: 8–35. [DOI: 10.1111/1750-3841.15998]
Nichols B., Quince C. (2016). Simera: modelling the PCR process to simulate realistic chimera formation. bioRxiv. [DOI: 10.1101/072447]
Nicolae C., Moga L., Bahaciu G., Marin M. (2017). Traceability system structure design for fish and fish products based on supply chain actors' needs. Food Control. 73: 189-200. [DOI: 10.1016/j.foodcont. 2016.10.032]
Noakes M.T., Brinkerhoff H., Laszlo A.H., Derrington I.M., Langford K.W., Mount J.W., Bowman J.L., Baker K., Doering K., Tickman B.I., Gundlach J.H. (2019). Increasing the accuracy of nanopore DNA sequencing using a time-varying cross membrane voltage. Nature Biotechnology. 37: 651–656. [DOI: 10.1038/s41587-019-0096-0]
Noor N.M., Azmi N.A.N., Hanifah N.A., Zamarudin Z. (2023). Navigating the halal food ingredients industry: exploring the present landscape. Halalpshere. 3: 32–43. [DOI: 10.31436/ hs.v3i2.80]
Nuraini H., Primasari A., Andreas E., Sumantri C. (2012). The use of Cytochrome b gene as a specific marker of the rat meat (Rattusnorvegicus) on meat and meat products. Media Peternakan. 35: 15–20. [DOI: 10.5398/medpet.2012.35.1.15]
Oehler J.B., Wright H., Stark Z., Mallett A., Schmitz U. (2023). The application of long-read sequencing in clinical settings. Human Genomics. 17: 92. [DOI: 10.1186/s40246-023-00522-3]
Oktavia L., Marwa T., Yulianita A. (2019). Analysis of factors affecting millennial consumers’ demand for halal bread products. MIR (Modernization Innovation Research). 10: 395–407. [DOI: 10.18184/2079-4665.2019.10.3.395-407]
Ounit R., Wanamaker S., Close T.J., Lonardi S. (2015). CLARK: fast and accurate classification of metagenomic and genomic sequences using discriminative k-mers. BMC Genomics. 16: 236. [DOI: 10.1186/s12864-015-1419-2]
Pandey P.K., Dhotre D.P., Dharne M.S., Khadse A.N., Hiremath U.I., Chaudhari R.D., Patole M.S., Shouche Y.S. (2007). Evaluation of mitochondrial 12S rRNA gene in the identification of Panterapardusfusca (Meyer, 1794) from field-collected scat samples in Western Ghats, Maharashtra, India. Current Science. 92: 1129-1133.
Pang Z., Zhou G., Ewald J., Chang L., Haçarız O., Basu N., Xia J. (2022). Using MetaboAnalyst 5.0 for LC–HRMS spectra processing, multi-omics integration, and covariate adjustment of global metabolomics data. Nature Protocols. 17: 1735-1761. [DOI: 10.1038/s41596-022-00715-4]
Pillay S., De Vos M., Derendinger B., Streicher E.M., Dolby T., Scott L.A., Steinhobel A.D., Warren R.M., Theron G. (2022). Non-actionable results, accuracy, and effect of first- and second-line line probe assays for diagnosing drug-resistant tuberculosis, including on smear-negative specimens, in a high-volume laboratory. Clinical Infectious Diseases. 76: e920–e929. [DOI: 10.1093/cid/ciac556]
Prakrongrak N., Boonsoong B., Monthatong M. (2023). Genetic diversity and phylogenetic analysis of mayfly Caenis (Insecta: Ephemeroptera) using Cytochrome C Oxidase I (COI) and 12s rRNA genes from Thailand. Biodiversitas Journal of Biological Diversity. 24: 1942-1952. [DOI: 10.13057/biodiv/ d240407]
Rahman M.M., Ahmad Z., Mustafa S. (2023). DNA-based platform for halal authentication and combat food threat. Journal of Halal Industry and Services. [DOI: 10.36877/jhis.a0000407]
Reuter J., Spacek D.V., Snyder M. (2015).High-throughput sequencing technologies. Molecular Cell. 58: 586–597. [DOI: 10.1016/j.molcel.2015.05.004]
Rhoads A., Au K.F. (2015). PacBio sequencing and its applications. Genomics, Proteomics and Bioinformatics. 13: 278–289. [DOI: 10.1016/j.gpb. 2015.08.002]
Ripp F., Krombholz C.F., Liu Y., Weber M., Schäfer A., Schmidt B., Köppel R., Hankeln T. (2014). All-Food-Seq (AFS): a quantifiable screen for species in biological samples by deep DNA sequencing. BMC Genomics. 15: 639. [DOI: 10.1186/1471-2164-15-639]
Rohman A., Orbayinah S., Hermawan A., Sudjadi S., Windarsih A., Handayani S. (2022). The development of real-time polymerase chain reaction for identification of beef meatball. Applied Food Research. 2: 100148. [DOI: 10.1016/j.afres.2022.100148]
Roy D., Akhter S., Sarker A., Hossain M., Lyzu C., Mohanta L., Islam D., Khan M. (2022). Tracing the pig and cattle origin in processed food and feed products targeting mitochondrial 12S rRNA gene. Journal of Food Quality and Hazards Control. 9: 163-171. [DOI: 10.18502/jfqhc.8.4.8256]
Sahilah A.M., Norhayati Y., Norrakiah A.S., Aminah A., Wan Aida W.M. (2011). Halal authentication of raw meats using PCR amplification of mitochondrial DNA. International Food Research Journal. 18: 1489-1491.
Sajali N., Wong S.C., Bakar S.A., Mokhtar N.F.K., Manaf Y.N.A., Yuswan M.H., Desa M.N.M. (2020). Analytical approaches of meat authentication in food. International Journal of Food Science and Technology. 56: 1535–1543. [DOI: 10.1111/ ijfs.14797]
Sanjaya U.H., Arabella R. (2023). Legal protection of consumer data on e-commerce platforms with cash on delivery (COD) systems. KnE Social Sciences. 8: 20-32. [DOI: 10.18502/kss.v8i1.10869]
Satam H., Joshi K., Mangrolia U., Waghoo S., Zaidi G., Rawool S., Thakare R.P., Banday S., Mishra A., Das G., Malonia S.K. (2023). Next-generation sequencing technology: current trends and advancements. Biology.12: 997. [DOI: 10.3390/biology12070997]
Schloss P.D., Gevers D., Westcott S.L. (2011). Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLOS ONE. 6: e27310. [DOI: 10.1371/journal.pone.0027310]
Sedlazeck F.J., Rescheneder P., Smolka M., Han F., Nattestad M., Von Haeseler A., Schatz M.C. (2018).Accurate detection of complex structural variations using single-molecule sequencing. Nature Methods. 15: 461–468. [DOI: 10.1038/s41592-018-0001-7]
Segata N., Waldron L., Ballarini A., Narasimhan V., Jousson O., Huttenhower C. (2012). Metagenomic microbial community profiling using unique clade-specific marker genes. Nature Methods. 9: 811–814. [DOI: 10.1038/nmeth.2066]
Shaharuddin F.Z., Norman A.A., Hamid S., Zakaria Z. (2025). Halal food industry in Malaysia: issues and challenges. Food Research. 9: 33–42. [DOI: 10.26656/fr.2017.9(s2).072]
Shen W., Song Z., Zhong X., Huang M., Shen D., Gao P., Qian X., Wang M., He X., Wang T., Li S., Song X. (2022). Sangerbox: a comprehensive, interaction‐friendly clinical bioinformatics analysis platform. iMeta. 1: 15. [DOI: 10.1002/imt2.15]
Shirazi S., Meyer R.S., Shapiro B. (2021). Revisiting the effect of PCR replication and sequencing depth on biodiversity metrics in environmental DNA metabarcoding. Ecology and Evolution. 11: 15766–15779. [DOI: 10.1002/ece3.8239]
Slatko B.E., Gardner A.F., Ausubel F.M. (2018).Overview of next-generation sequencing technologies. Current Protocols in Molecular Biology. 122: e59. [DOI: 10.1002/cpmb.59]
Sun K., Liu Y., Zhou X., Yin C., Zhang P., Yang Q., Mao L., Shentu X., Yu X. (2022). Nanopore sequencing technology and its application in plant virus diagnostics. Frontiers in Microbiology. 13: 939666. [DOI: 10.3389/fmicb.2022.939666]
Taber K.S. (2017). The use of Cronbach’s Alpha when developing and reporting research instruments in science education. Research in Science Education. 48: 1273–1296. [DOI: 10.1007/s11165-016-9602-2]
Talib M.S.A., Zulfakar M.H. (2023). Sustainable halal food supply chain management in a small rentier halal market. Arab Gulf Journal of Scientific Research. 41: 61-72. [DOI: 10.1108/agjsr-11-2022-0251]
Teng J.L.L., Yeung M.L., Chan E., Jia L., Lin C.H., Huang Y., Tse H., Wong S.S.Y., Sham P.C., Lau S.K.P., Woo P.C.Y. (2017). PacBio but not Illumina technology can achieve fast, accurate and
complete closure of the high GC, complex Burkholderiapseudomallei two-chromosome genome. Frontiers in Microbiology. 8: 1448. [DOI: 10.3389/fmicb.2017.01448]
Thomas G.W.C., Hahn M.W. (2019). Referee: reference assembly quality scores. Genome Biology and Evolution. 11: 1483–1486. [DOI: 10.1093/ gbe/evz088]
Tyler A.D., Mataseje L., Urfano C., Schmidt L., Antonation K., Mulvey M.R., Corbett C.R. (2018). Evaluation of Oxford Nanopore’sMinION sequencing device for microbial whole genome sequencing applications. Scientific Reports. 8: 10931. [DOI: 10.1038/s41598-018-29334-5]
Udaondo Z., Sittikankaew K., Uengwetwanit T., Wongsurawat T., Sonthirod C., Jenjaroenpun P., Pootakham W., Karoonuthaisiri N., Nookaew I. (2021). Comparative analysis of PacBio and Oxford Nanopore sequencing technologies for transcriptomic landscape identification of Penaeusmonodon. Life. 11: 862. [DOI: 10.3390/life11080862]
Uliano-Silva M., Ferreira J.G.R.N., Krasheninnikova K., Blaxter M., Mieszkowska N., Hall N., Holland P., Durbin R., Richards T., Kersey P., Hollingsworth P., Wilson W., Twyford A., Gaya E., Lawniczak M., Lewis O., Broad G., Martin F., Hart M., McCarthy S.A. (2023). MitoHiFi: a python pipeline for mitochondrial genome assembly from PacBio high fidelity reads. BMC Bioinformatics. 24: 288. [DOI: 10.1186/s12859-023-05385-y]
Van Der Spiegel M., Van Der Fels-Klerx H., Sterrenburg P., Van Ruth S.M., Scholtens-Toma I., Kok E. (2012). Halal assurance in food supply chains: verification of halal certificates using audits and laboratory analysis. Trends in Food Science and Technology. 27: 109–119. [DOI: 10.1016/j.tifs.2012.04.005]
Venkatas J., Adeleke M.A. (2019). Emerging threat of Eimeria operational taxonomic units (OTUs) on poultry production. Parasitology. 146: 1615–1619. [DOI: 10.1017/s0031182019001100]
Wang H., Chen Y., Wang L., Liu Q., Yang S., Wang C. (2023). Advancing herbal medicine: enhancing product quality and safety through robust quality control practices. Frontiers in Pharmacology. 14: 1265178. [DOI: 10.3389/fphar.2023.1263813]
Wang X., Cai Y., Sun Y., Knight R., Mai V. (2011). Secondary structure information does not improve OTU assignment for partial 16S rRNA sequences. The ISME Journal. 6: 1277–1280. [DOI: 10.1038/ismej.2011.187]
Wasswa F.B., Kassaza K., Nielsen K., Bazira J. (2022). MinION whole-genome sequencing in resource-limited settings: challenges and opportunities. Current Clinical Microbiology Reports. 9: 52–59. [DOI: 10.1007/s40588-022-00183-1]
Wenger A., Peluso P., Rowell W., Chang P.C., Hall R., Concepcion G., Hunkapiller M. (2019). Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nature Biotechnology. 37: 1155–1162. [DOI: 10.1038/s41587-019-0217-9]
Wick R.R., Judd L.M., Holt K.E. (2023). Assembling the perfect bacterial genome using Oxford Nanopore and Illumina sequencing. PLOS Computational Biology. 19: e1010905. [DOI: 10.1371/journal.pcbi.1010905]
Woltyńska A., Gawor J., Olech M.A., Górniak D., Grzesiak J. (2023). Bacterial communities of Antarctic lichens explored by gDNA and cDNA 16S rRNA gene amplicon sequencing. FEMS Microbiology Ecology. 99: fiad015. [DOI: 10.1093/femsec/fiad015]
Woo C., Kumari P., Eo K., Lee W., Kimura J., Yamamoto N. (2023). Combining vertebrate mitochondrial 12S rRNA gene sequencing and shotgun metagenomic sequencing to investigate the diet of the leopard cat (Prionailurusbengalensis) in Korea. PLOS ONE. 18: e0278941. [DOI: 10.1371/journal.pone.0278941]
Wood D.E., Lu J., Langmead B. (2019). Improved metagenomic analysis with Kraken 2. Genome Biology. 20: 257. [DOI: 10.1186/s13059-019-1891-0]
Yakin A.U. (2021). Halal certification, standards, and their ramifications in Belgium. Islamic Studies Journal. 27: 45-59. [DOI: 10.1163/9789004459236_ 008]
Yang Y., Xie B., Yan J. (2014). Application of next-generation sequencing technology in forensic science. Genomics, Proteomics and Bioinformatics. 12: 190–197. [DOI: 10.1016/j.gpb.2014.09.001]
Yang L., Tan Z., Wang D., Xue L. Guan M., Huang T., Li R. (2014). Species identification through mitochondrial rRNA genetic analysis. Scientific Reports. 4: 4089. [DOI: 10.1038/srep04089]
Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., Madden T.L. (2012). Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics.13: 134. [DOI: 10.1186/ 1471-2105-13-134]
Zhang Y., Qu Q., Mingzhen R., Nana Z., Zhao Y., Tao F. (2020). Simultaneous identification of animal-derived components in meats using high-throughput sequencing in combination with a custom-built mitochondrial genome database. Scientific Reports.10: 8965. [DOI: 10.1038/s41598-020-65724-4]
Zhou W., Liu X., Lv M., Shi Y., Zhang L. (2023). The recognition mode between hsRBFA and mitoribosome 12S rRNA during mitoribosomal biogenesis. Nucleic Acids Research. 51: 1353–1363. [DOI: 10.1093/ nar/gkac1234]
Zou A., Phan L., Chen S., Campbell J., Guo P., Ren R., Hendrycks D. (2023). Representation engineering: a top-down approach to AI transparency. arXiv. [DOI: 10.48550/arXiv.2301.01912]
Food and safety in halal context
The introduction of innovative molecular approaches for halal authentication highlights their importance in ensuring food safety. This approach provides a holistic method to make sure that halal foods are wholesome and safe, at the expense of religious dogma and firm science. At the same time, the contents of food products should be validated to determine whether they are authentic and whether they originate from the claimed sources (Daniel et al. 2022). The present study emphasises mitochondrial 12S rRNA sequencing (El-Nabi et al., 2021) as an approach that strikes at the heart of safety concerns associated with both intentional or unintentional contamination of halal foods with haram components.
Visual or manual inspection methodologies are incapable of guaranteeing food safety, and as such, molecular methodologies are crucially necessary in food processing. The risk of cross-contamination or false labelling is eradicated, by detecting even the slightest trace of a non-halal ingredient. For consumers, this is even more significant, especially those with dietary restrictions or allergies, which make it critical to avoid harmful substances (Hidayat et al., 2023). Food is manufactured and spread around the world, and this globalised nature creates huge risks of contaminants and safety breaches. Ensuring that ingredients, manufacturing processes, and handling conform to both safety standards and religious beliefs is particularly challenging in the realm of halal food safety. With the combinations of NGS and bioinformatics methods, a complete identification of contamination of the whole supply chain is presented (Fu et al., 2012).
The techniques described here are not only used to ascertain the truth of a species but also to characterise microbiological contaminants, which guarantee the good of food products from horrible infections (Delahoy et al., 2023). This dual molecular technology protects against foodborne infections, which are a serious global public health concern, while also complying with halal certification (Krisna and Yusuf, 2023). Hence, cleanliness and hygiene are key elements of halal food safety, built on the foundation of Tayyib, which emphasises purity and wholesomeness (Aksin and Aini, 2023).
Best practices to ensure halal traceability
This review represents significant progress in the halal supply chain, utilising technology, accountability, and transparency. Indeed, traceability systems are essential for maintaining food safety, enabling stakeholders to trace ingredients from farm to fork and guarantee that all relevant parties in the supply chain comply with regulation (Bosona and Gebresenbet, 2023). Technology provides an additional level of transparency that makes it possible to detect and respond quickly to contamination events and safety violations.
This then results in the integration of recent scientific findings with accepted religious guidelines and makes meaningful contributions toward the discussion within the halal paradigm. Their suggested approaches pave the way for ethical and technological inclusion in the halal space while successfully addressing the intricacies of modern supply chains (Cobbe et al., 2023). Fighting these battles will require innovation and embracing creative solutions in doing so is critical. This will serve as a cornerstone for preserving the integrity of halal-certified products, as it guarantees their safety and reliability as global consumption continues to rise (Oktavia et al., 2019).
Future perspectives and applications of halal products
The above-mentioned approach demonstrates how full utilisation of integrated state-of-the-art technologies such as multi-omics approaches is essential to enhance compliance, quality assurance, and transparency standards in the halal industry (Pang et al., 2022). This vision serves as a reflection of the growing worldwide focus on halal products, while also highlighting the need for technology to meet consumer demands and industry standards (Ezeaku et al., 2021).
It is a compelling technical approach in that it links leading-edge scientific innovations to practical solutions. For instance, the advent of portable sequencing devices as well as in-field, automated data analysis tools provides a compelling pathway for efficiency gains and lower barriers to authentication validation to stakeholders (Hayes et al., 2021). In addition, the request for common sequencing technologies and standardised analysis pipelines demonstrates a deeper understanding of the challenges to harmonisation of methods and maintaining comparability across many disparate regions and laboratories (Algan, 2023).
A particularly compelling link exists between halal authenticity and food safety. Based on the technicalities of 12S rRNA sequencing, which resonate with the general theme of well-being in a consumer era, the success rests on the identification of commonalities around hygiene to prevent contamination and assurance of computation quality (Kumari et al., 2019). The complementarity of both aspects signifies the need for a joint application of halal compliance and food safety, strengthening the underlying rationale of using modern technologies for these aims.
The ensuing discussion should include the practical implementation of these metagenomic and DNA approaches in halal testing pipelines (Cantalapiedra et al. 2021). Similarly, accounting for complicating factors, such as the cost and plausibility of incorporating technologies like portable sequencing devices would provide a clearer picture (Hayes et al. 2021). Another way to improve reliability would be by directly correlating ethics with issues arising from data privacy in consumer platforms technology (Gallardo et al., 2023; Sanjaya and Arabella, 2023).
Moreover, including practical examples and/or case studies from other industries with similar traceability needs (e.g., kosher certification and broader food safety needs) would further help readers understand the practicability of the proposals (Nicolae et al., 2017). Addressing specific omissions in existing halal certification across regions may also provide a strong justification for the proposed standardisation efforts (Yakin, 2021). Lastly, capturing the interest of readers by focusing on the sustainability aspect of these technologies for instance, their economic and environmental advantages, such as the reduction of waste or the optimisation of resources utilised in halal production, would further strengthen the discussion (Allam et al., 2022; Jiang, 2023). One of its significant strengths is its consumer-driven, privacy-preserving solutions (e.g., platforms that allow consumers to directly access the result of the authenticity test) (Muralidharan et al., 2023). By emphasising transparency, these solutions contribute to enhancing consumer trust, an important component in the global halal space (Talib and Zulfakar, 2023). However, some thoughts on how these platforms could be created, implemented, and maintained (especially in low-resource settings), would provide a practical dimension to this claim (Mathew, 2023).
Conclusion
The industrial-scale DNA sequencing technologies discussed in this article support halal authentication, thereby enabling stakeholders and consumers to achieve greater transparency, compliance, and trust. To address concerns regarding authenticity and traceability issues in halal, this study utilised modern sequencing techniques as a credible means for validating halal product integrity. It also serves to reinforce the moral aspects within the supply chain as well as to encourage the growth and long-term viability of the halal industry. Consequently, the industry is increasingly adopting advanced methods such as DNA sequencing to provide consumers with verified and up-to-date verification of a product's halal status. This is important in combating fraud, mislabelling, and contamination, thereby supporting the religious and cultural preferences of consumers.
In addition, the global acceleration of commerce and communication has strengthened the need for halal standardisation of halal regulations on an international scale. Industrial-scale sequencing technology supports this by offering companies a simple solution to cross-navigate the heterogeneous regulatory landscape in the jurisdictions through platforms designed to standardise and unify the halal certification process. The broader long-term benefits of this approach certainly outweigh its cost. However, the technical sophistication does come with its own costs. Nevertheless, the technology complements consumer trust, reinforces brand equity, and aligns with the moral and religious beliefs of consumers, thus nurturing the sustainable growth of the halal industry. Moreover, sequencing technology provides transparency that caters to the increasing demand for information and ethical practices in today’s food system.
Author contributions
J.J. and M.A.S. designed the study; J.J., M.A.S, C.L.S.L. and S.M. collected the data J.J., M.A.S, C.L.S.L. and S.M. analyzed the data and wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgements
The authors are grateful to the team members for their contributions of ideas, knowledge, and financial support, especially Constance and Symeon from the Department of Anesthesiology and Intensive Care, Faculty of Medicine and Health Sciences, Kota Kinabalu, Sabah, Malaysia, and from the School of Business, Swinburne University of Technology, Kuching, Sarawak.
Conflicts of interest
The authors declare that there is no conflict of interest.
Funding
This research received no specific grant from any funding agency in the public, commercial, or non-profit sectors.
Ethical consideration
Not applicable.
References
Absmeier E., Chandrasekaran V., O’Reilly F.J., Stowell J.a.W., Rappsilber J., Passmore L.A. (2023). Specific recognition and ubiquitination of translating ribosomes by mammalian CCR4–NOT. Nature Structural and Molecular Biology. 30: 1314–1322. [DOI: 10.1038/s41594-023-01075-8]
Ahsan M.U., Liu Q., Perdomo J.E., Fang L., Wang K. (2023). A survey of algorithms for the detection of genomic structural variants from long-read sequencing data. Nature Methods. 20: 1143–1158. [DOI: 10.1038/s41592-023-01932-w]
Aini S.R., Rohman A., Mulyanto M., Erwanto Y., Ansar A., Handayani B.R., Irnawati I. (2023). The metabolomics approach used for halal authentication analysis of food and pharmaceutical products: a review. Food Research. 7: 180–187.
Aksamentov I., Roemer C., Hodcroft E.B., Neher R.A. (2021). Nextclade: clade assignment, mutation calling and quality control for viral genomes. Journal of Open Source Software. 6: 3773. [DOI: 10.21105/joss.03773]
Aksin N., Aini F.N.Q. (2023). Halal Ṭayyib syaria’ in understanding the needs of consumer protection. Jurnal Meta-Yuridis. 6: 1–13. [DOI: 10.26877/m-y.v6i2.15560]
Algan F.M. (2023). Global standardization for a sustainable future. Current Research in Social Sciences. 9: 30-40. [DOI: 10.30613/ curesosc.1173926]
Allam Z., Bibri S.E., Chabaud D., Moreno C. (2022). The theoretical, practical, and technological foundations of the 15-minute city model: proximity and its environmental, social, and economic benefits for sustainability. Energies. 15: 6042. [DOI: 10.3390/ en15166042]
Amarasinghe S.L., Su S., Dong X., Zappia L., Ritchie M.E., Gouil Q. (2020). Opportunities and challenges in long-read sequencing data analysis. Genome Biology. 21: 30. [DOI: 10.1186/s13059-020-1935-5]
Amer M. (2024). Halal standards’ implementation in Palestinian food sector: its drivers and impact on performance. Arab Gulf Journal of Scientific Research. 42: 2–29. [DOI: 10.1108/agjsr-09-2022-0168]
Aziz N., Bakry N., Habibi Mz M., Siddiq Armia M. (2023). The paradigm of modern food products and its relevance with the concept of food in the Quran. Heliyon. 9: e21358. [DOI: 10.1016/j. heliyon.2023.e21358]
Bannor R.K., Arthur K.E., Oppong D., Oppong-Kyeremeh H. (2023). A comprehensive systematic review and bibliometric analysis of food fraud from a global perspective. Journal of Agriculture and Food Research. 14: 100686. [DOI: 10.1016/j.jafr. 2023.100686]
Bianconi I., Aschbacher R., Pagani E. (2023). Current uses and future perspectives of genomic technologies in clinical microbiology. Antibiotics. 12: 1580. [DOI: 10.3390/antibiotics12111580]
Bohmann K., Elbrecht V., Carøe C., Bista I., Leese F., Bunce M., Yu D.W., Seymour M., Dumbrell A.J., Creer S. (2022). Strategies for sample labelling and library preparation in DNA metabarcoding studies. Molecular Ecology Resources. 22: 1231–1246. [DOI: 10.1111/1755-0998.13512]
Bolger A.M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30: 2114–2120. [DOI: 10.1093/bioinformatics/btu170]
Borodinov A., Manoilov V., Zarutsky I., Petrov A., Kurochkin V., Saraev A. (2022). Machinelearning in base-calling for next-generation sequencing methods. Informatics and Automation. 21: 572–603. [DOI: 10.15622/ia.21.3.5]
Bosona T., Gebresenbet G. (2023). The role of blockchain technology in promoting traceability systems in agri-food production and supply chains. Sensors. 23: 5342. [DOI: 10.3390/s23115342]
Boyrusbianto F.T., Utama D.T., Khikmawati I., Fadhila A., Volkandari S.D., Abdurrahman Z.H., Pramono A., Cahyadi M. (2023). A simplex and multiplex PCR assay for simultaneous detection of beef, pork, and chicken meat in sausages based on mitochondrial DNA Cytochrome oxidase sub-unit I. Food Research. 7: 188–193. [DOI: 10.26656/fr.2017.7(3).084]
Cahyadi M., Wibowo T., Pramono A., Abdurrahman Z.H. (2020). A novel multiplex-PCR assay to detect three non-halal meats contained in meatballs using mitochondrial 12S rRNA gene. Food Science of Animal Resources. 40: 628–635. [DOI: 10.5851/kosfa.2020.e40]
Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nature Methods. 13: 581–583. [DOI: 10.1038/ nmeth.3869]
Cantalapiedra C.P., Hernández-Plaza A., Letunic I., Bork P., Huerta-Cepas J. (2021). eggNOG-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Molecular Biology and Evolution. 38: 5825-5829. [DOI: 10.1093/molbev/msab293]
Cao M.D., Ganesamoorthy D., Cooper M.A., Coin L.J.M. (2015). Realtime analysis and visualization of MinION sequencing data with npReader. Bioinformatics. 32: 764–766. [DOI: 10.1093/bioinformatics/btv658]
Chan A.H.E., Saralamba N., Saralamba S., Ruangsittichai J., Thaenkham U. (2022). The potential use of mitochondrial ribosomal genes (12S and 16S) in DNA barcoding and phylogenetic analysis of trematodes. BMC Genomics. 23: 104. [DOI: 10.1186/s12864-022-08302-4]
Chang J.J.M., Ip Y.C.A., Bauman A.G., Huang D. (2020). MINION-in-ARMS: nanopore sequencing to expedite barcoding of specimen-rich macrofaunal samples from autonomous reef monitoring structures. Frontiers in Marine Science. 7: 448. [DOI: 10.3389/fmars.2020. 00448]
Chen C., Wu Y., Li J., Wang X., Zeng Z., Xu J., Liu Y., Feng J., Chen H., He Y., Xia R. (2023). TBtools-II: a "one for all, all for one" bioinformatics platform for biological big-data mining. Molecular Plant. 16: 1733-1742. [DOI: 10.1016/j.molp.2023.09.010]
Chen S., Zhou Y., Chen Y., Gu J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34: i884–i890. [DOI: 10.1093/bioinformatics/bty560]
Cobbe J., Veale M., Singh J. (2023). Understanding accountability in algorithmic supply chains. Proceedings of the 2023 ACM Conference on Fairness, Accountability, and Transparency. 1186–1197. [DOI: 10.1145/3593013.3594073]
Daniel A.I., Fadaka A.O., Gokul A., Bakare O.O., Aina O., Fisher S., Burt A.F., Mavumengwana V., Keyster M., Klein A. (2022). Biofertilizer: the future of food security and food safety. Microorganisms. 10: 1220. [DOI: 10.3390/microorganisms10061220]
De Coster W., Weissensteiner M.H., Sedlazeck F.J. (2021). Towards population-scale long-read sequencing. Nature Reviews Genetics. 22: 572–587. [DOI: 10.1038/s41576-021-00367-3]
Delahoy M.J., Shah H.J., Weller D.L., Ray L.C., Smith K., McGuire S., Trevejo R.T., Walter E.S., Wymore K., Rissman T., McMillian M., Lathrop S., et al. (2023). Preliminary incidence and trends of infections caused by pathogens transmitted commonly through food — foodborne diseases active surveillance network, 10 U.S. sites, 2022. Morbidity and Mortality Weekly Report. 72: 701–706. [DOI: 10.15585/ mmwr.mm7226a1]
Dubois B., Debode F., Hautier L., Hulin J., Martin G.S., Delvaux A., Janssen E., Mingeot D. (2022). A detailed workflow to develop QIIME2-formatted reference databases for taxonomic analysis of DNA metabarcoding data. BMC Genomic Data. 23: 53. [DOI: 10.1186/s12863-022-01045-w]
Dunham J.P., Friesen M.L. (2013). A cost-effective method for high-throughput construction of Illumina sequencing libraries. Cold Spring Harbor Protocols. 2013: 820-834. [DOI: 10.1101/pdb.prot074187]
El-Nabi S.E.H., Hussein D., Khallafa A.G. (2021). Molecular detection of food fraud targeting mitochondrial 12S rRNA gene sequencing. Journal of Bioscience and Applied Research. 7: 17–22. [DOI: 10.21608/jbaar.2021.167091]
Ezeaku H.C., Asongu S.A., Nnanna J. (2021). Volatility of international commodity prices in times of COVID-19: effects of oil supply and global demand shocks. The Extractive Industries and Society. 8: 257-270. [DOI: 10.1016/j.exis.2020.12.013]
Ferreira T., Rodriguez S. (2024). Mitochondrial DNA: inherent complexities relevant to genetic analyses. Genes. 15: 617. [DOI: 10.3390/ genes15050617]
Fosso B., Santamaria M., Marzano M., Alonso-Alemany D., Valiente G., Donvito G., Monaco A., Notarangelo P., Pesole G. (2015). BioMaS: a modular pipeline for bioinformatic analysis of metagenomic amplicons. BMC Bioinformatics. 16: 203. [DOI: 10.1186/s12859-015-0595-z]
Fu L., Niu B., Zhu Z., Wu S., Li W. (2012). CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 28: 3150–3152. [DOI: 10.1093/bioinformatics/bts565]
Galal-Khallaf A. (2021). Multiplex PCR and 12S rRNA gene sequencing for detection of meat adulteration: a case study in the Egyptian markets. Gene. 764: 145062. [DOI: 10.1016/j.gene.2020.145062]
Gallardo A., Choy C., Juneja J., Bozkir E., Cobb C., Bauer L., Cranor L. (2023). Speculative privacy concerns about AR glasses data collection. Proceedings on Privacy Enhancing Technologies. 2023: 416-435. [DOI: 10.56553/ popets-2023-0117]
Garinet S., Laurent-Puig P., Blons H., Oudart J. (2018). Current and future molecular testing in NSCLC: what can we expect from new sequencing technologies? Journal of Clinical Medicine. 7: 144. [DOI: 10.3390/jcm7060144]
Gupta N., Verma V. (2019). Next-generation sequencing and its application: empowering in public health beyond reality. In: Arora P. (Editor). Microbial technology for the welfare of society. Springer, Singapore. pp: 313–341. [DOI: 10.1007/978-981-13-8844-6_15]
Hassan N., Ahmad T., Zain N.M., Ashaari A. (2020). A novel chemometrics method for halal authentication of gelatin in food products. SainsMalaysiana. 49: 2083–2089. [DOI: 10.17576/jsm-2020-4907-08]
Hayes B., Nguyen L.T., Forutan M., Engle B.N., Lamb H.J., Copley J.P., Randhawa I.A.S., Ross E.M. (2021). An epigenetic aging clock for cattle using portable sequencing technology. Frontiers in Genetics. 18: 760450. [DOI: 10.3389/fgene. 2021.760450]
Haynes E., Jiménez E., Pardo M.Á., Helyar S.J. (2019). The future of NGS (Next Generation Sequencing) analysis in testing food authenticity. Food Control. 101: 134–143. [DOI: 10.1016/j.foodcont. 2019.02. 010]
Hernández V.P., Guzmán M.H. (2023). Microorganisms identification in environmental samples: bioinformatic analysis of 16S rRNA gene with QIIME2. Ciencia Latina Revista Científica Multidisciplinar. 7: 8074–8102. [DOI: 10.37811/cl_rcm.v7i5.8382]
Hidayat H.H., Wijayanti N., Situmorang S.C.D.U.B. (2023). Materials clustering of Soto Sokaraja as an effort to accelerate halal certification. IOP Conference Series: Earth and Environmental Science. 1177: 012036. [DOI: 10.1088/1755-1315/1177/ 1/012036]
Hook P.W., Timp W. (2023). Beyond assembly: the increasing flexibility of single-molecule sequencing technology. Nature Reviews Genetics. 24: 627–641. [DOI: 10.1038/s41576-023-00600-1]
Jani N.J., Chin N.K.L., Mustapha N.Z.A. (2023). Whole genome sequence analysis of a Mycobacterium iranicum, a newly identified non-tuberculosis mycobacteria, strain SBH312 isolated in Sabah, Malaysia. Borneo Journal of Medical Sciences. 17: 52–56. [DOI: 10.51200/bjms.v17i1.4291]
Jaswir I., Mirghani M.E.S., Salleh H.M., Ramli N., Octavianti F., Hendri R. (2016). An overview of the current analytical methods for halal testing. Current issues in halal food analysis. Springer, Singapore. pp: 291–300. [DOI: 10.1007/978-981-10-1452-9_27]
Jiang L. (2023). Environmental benefits of green buildings with BIM technology. Ecological Chemistry and Engineering. 30: 191-199. [DOI: 10.1515/eces-2023-0003]
Jovic D., Liang X., Zeng H., Lin L., Xu F., Luo Y. (2022). Single‐cell RNA sequencing technologies and applications: a brief overview. Clinical and Translational Medicine. 12: 21-35. [DOI: 10.1002/ ctm2.983]
Karst S.M., Ziels R.M., Kirkegaard R., Sørensen E.A., McDonald D., Zhu Q., Knight R., Albertsen M. (2021). High-accuracy long-read amplicon sequences using unique molecular identifiers with Nanopore or PacBio sequencing. Nature Methods. 18: 165-169. [DOI: 10.1038/s41592-020-01041-y]
Katoh K., Rozewicki J., Yamada K.D. (2017). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics. 20: 1160–1166. [DOI: 10.1093/bib/bbx108]
Kaveh S., Hashemi S.M.B., Abedi E., Amiri M.J., Conte F.L. (2023). Bio-preservation of meat and fermented meat products by lactic acid bacteria strains and their antibacterial metabolites. Sustainability. 15: 10154. [DOI: 10.3390/su151310154]
Kayama K., Kanno M., Chisaki N., Tanaka M., Yao R., Hanazono K., Camer G.A., Endoh D. (2021). Prediction of PCR amplification from primer and template sequences using recurrent neural network. Scientific Reports. 11: 7493. [DOI: 10.1038/s41598-021-86357-1]
King J., Harder T., Beer M., Pohlmann A. (2020). Rapid multiplex MinION nanopore sequencing workflow for Influenza A viruses. BMC Infectious Diseases. 20: 648. [DOI: 10.1186/s12879-020-05367-y]
Kozich J.J., Westcott S.L., Baxter N.T., Highlander S.K., Schloss P.D. (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MISEQ Illumina sequencing platform. Applied and Environmental Microbiology. 79: 5112–5120. [DOI: 10.1128/aem. 01043-13]
Krisna R., Yusuf M. (2023). Halal ecosystem improvement study reviewed of halal product regulations. International Journal of Research and Review. 10: 339–353. [DOI: 10.52403/ijrr.20230243]
Kumari P., Dong K., Eo K., Lee W., Kimura J., Yamamoto N. (2019). DNA metabarcoding-based diet survey for the Eurasian otter (Lutralutra): Development of a Eurasian otter-specific blocking oligonucleotide for 12S rRNA gene sequencing for vertebrates. PLOS ONE. 14: e0226253. [DOI: 10.1371/journal.pone.0222966]
Liu H., Nie J., Liu Y., Wadood S.A., Rogers K.M., Yuan Y., Gan R. (2023). A review of recent compound-specific isotope analysis studies applied to food authentication. Food Chemistry. 415: 135791. [DOI: 10.1016/j.foodchem.2023.135791]
Liu J., Seetharam A.S., Chougule K., Ou S., Swentowsky K.W., Gent J.I., Llaca V., Woodhouse M.R., Manchanda N., Presting G.G., Kudrna D.A., Alabady M., Hirsch C.N., Fengler K.A., Ware D., Michael T.P., Hufford M.B., Dawe R.K. (2020). Gapless assembly of maize chromosomes using long-read technologies. Genome Biology. 21: 121. [DOI: 10.1186/s13059-020-02029-9]
Liu X., Liu Z., Cheng Y., Wu H., Shen W., Liu Y., Feng Q., Yang J. (2021). Application of next-generation sequencing technology based on single gene locus in species identification of mixed meat products. Journal of Food Quality. 2021: 4512536. [DOI: 10. 1155/2021/4512536]
Lu H., Giordano F., Ning Z. (2016). Oxford NanoporeMinION sequencing and genome assembly. Genomics, Proteomics and Bioinformatics. 14: 265–279. [DOI: 10.1016/j.gpb.2016.05.004]
Lubis H., Mohd-Naim N.F., Alizul N.N., Ahmed M.U. (2016). From market to food plate: current trusted technology and innovations in halal food analysis. Trends in Food Science and Technology. 58: 55–68. [DOI: 10.1016/j.tifs.2016.10.024]
Lücking R., Aime M.C., Robbertse B., Miller A., Ariyawansa H., Aoki T., Papp V., Robert V., Schoch C. (2020). Unambiguous identification of fungi: where do we stand and how accurate and precise is fungal DNA barcoding? IMA Fungus. 11: 14. [DOI: 10.1186/s43008-020-00033-9]
Luo J., Yan D., Zhang D., Han Y., Dong X., Yang Y., Deng K., Xiao X. (2011). Application of 12S rRNA gene for the identification of animal-derived drugs. Journal of Pharmacy and Pharmaceutical Sciences. 14: 358–367. [DOI: 10.18433/j3n017]
Ma X., Shao Y., Tian L., Flasch D.A., Mulder H.L., Edmonson M.N., Liu Y., Chen X., Newman S., Nakitandwe J., Li Y., Li B., Shen S., Wang Z., Shurtleff S., Robison L.L., Levy S., Easton J., Zhang J. (2019). Analysis of error profiles in deep next-generation sequencing data. Genome Biology. 20: 50. [DOI: 10.1186/s13059-019-1659-6]
Malaysia’s Halal Food Opportunities (MIDA). (2022). Malaysia’s halal food opportunities - carving out the lion’s share in a USD3 trillion global market. URL: https://www.mida.gov.my/malaysias-halal-food-opportunitiescarving-out-the-lions-share-in-a-usd3-trillion-global-market/. Accessed on 5 January 2025.
Mathew J.L. (2023). Efficiency, economy, and excellence: experimental exploration of evidence-based guideline development in resource-constrained settings. Indian Journal of Pediatrics. 90: 700-707. [DOI: 10.1007/s12098-023-03785-1]
Matsunaga T., Chikuni K., Tanabe R., Muroya S., Shibata K., Yamada J., Shinmura Y. (1999). A quick and simple method for the identification of meat species and meat products by PCR assay. Meat Science. 51: 143–148. [DOI: 10.1016/s0309-1740(98)00112-0]
Menzel P., Ng K.L., Krogh A. (2016). Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nature Communications. 7: 11257. [DOI: 10.1038/ncomms11257]
Meyer M., Kircher M. (2010). Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harbor Protocols. 2010: pdb.prot5448. [DOI: 10.1101/pdb.prot5448]
Mitchell A.L., Almeida A., Beracochea M., Boland M., Burgin J., Cochrane G., Crusoe M.R., Kale V., Potter S.C., Richardson L.J., Sakharova E., Scheremetjew M., et al. (2020). MGnify: the microbiome analysis resource in 2020. Nucleic Acids Research. 48: D570–D578. [DOI: 10.1093/nar/gkz1035]
Moorthie S., Mattocks C., Wright C.F. (2011). Review of massively parallel DNA sequencing technologies. The HUGO Journal. 5: 1–12. [DOI: 10.1007/s11568-011-9156-3]
Montiel-Sosa J.F., Ruiz-Pesini E., Montoya J., Roncalés P., López-Pérez M.J., Pérez-Martos A. (2000). Direct and highly species-specific detection of pork meat and fat in meat products by PCR amplification of mitochondrial DNA. Journal of Agricultural and Food Chemistry. 48: 2829–2832. [DOI: 10.1021/jf9907438]
Monzel A.S., Enríquez J.A., Picard M. (2023). Multifaceted mitochondria: moving mitochondrial science beyond function and dysfunction. Nature Metabolism. 5: 546–562. [DOI: 10.1038/s42255-023-00783-1]
Muflihah, Hardianto A., Kusumaningtyas P., Prabowo S., Hartati Y.W. (2023). DNA-based detection of pork content in food. Heliyon. 9: e14418. [DOI: 10.1016/j.heliyon.2023.e14418]
Muralidharan S., Yoo B., Ko H. (2023). Decentralized ME-centric framework—a futuristic architecture for consumer IoT. IEEE Consumer Electronics Magazine. 12: 39-50. [DOI: 10.1109/ MCE. 2023.3152809]
Murugaiah C., Noor Z.M., Mastakim M., Bilung L.M., Selamat J., Radu S. (2009). Meat species identification and halal authentication analysis using mitochondrial DNA. Meat Science. 83: 57–61. [DOI: 10.1016/j.meatsci.2009.03.015]
Nasir S., Zulfakar M.H., Rahmat A. (2021). Determinants of preferred export logistics gateway in Malaysia halal product industry. International Journal of Academic Research in Accounting, Finance and Management Sciences. 11: 60-69. [DOI: 10.6007/IJARAFMS/v11-i4/11823]
Neequaye G., Agyekum K., Botchway E. (2017). Market analysis of halal food consumption. International Journal of Food Science. 52: 112-124. [DOI: 10.1002/ijfs.2017.112]
Ng P.C., Ruslan N.A.S.A., Chin L.X., Ahmad M., Hanifah S.A., Abdullah Z., Khor S.M. (2021). Recent advances in halal food authentication: challenges and strategies. Journal of Food Science. 87: 8–35. [DOI: 10.1111/1750-3841.15998]
Nichols B., Quince C. (2016). Simera: modelling the PCR process to simulate realistic chimera formation. bioRxiv. [DOI: 10.1101/072447]
Nicolae C., Moga L., Bahaciu G., Marin M. (2017). Traceability system structure design for fish and fish products based on supply chain actors' needs. Food Control. 73: 189-200. [DOI: 10.1016/j.foodcont. 2016.10.032]
Noakes M.T., Brinkerhoff H., Laszlo A.H., Derrington I.M., Langford K.W., Mount J.W., Bowman J.L., Baker K., Doering K., Tickman B.I., Gundlach J.H. (2019). Increasing the accuracy of nanopore DNA sequencing using a time-varying cross membrane voltage. Nature Biotechnology. 37: 651–656. [DOI: 10.1038/s41587-019-0096-0]
Noor N.M., Azmi N.A.N., Hanifah N.A., Zamarudin Z. (2023). Navigating the halal food ingredients industry: exploring the present landscape. Halalpshere. 3: 32–43. [DOI: 10.31436/ hs.v3i2.80]
Nuraini H., Primasari A., Andreas E., Sumantri C. (2012). The use of Cytochrome b gene as a specific marker of the rat meat (Rattusnorvegicus) on meat and meat products. Media Peternakan. 35: 15–20. [DOI: 10.5398/medpet.2012.35.1.15]
Oehler J.B., Wright H., Stark Z., Mallett A., Schmitz U. (2023). The application of long-read sequencing in clinical settings. Human Genomics. 17: 92. [DOI: 10.1186/s40246-023-00522-3]
Oktavia L., Marwa T., Yulianita A. (2019). Analysis of factors affecting millennial consumers’ demand for halal bread products. MIR (Modernization Innovation Research). 10: 395–407. [DOI: 10.18184/2079-4665.2019.10.3.395-407]
Ounit R., Wanamaker S., Close T.J., Lonardi S. (2015). CLARK: fast and accurate classification of metagenomic and genomic sequences using discriminative k-mers. BMC Genomics. 16: 236. [DOI: 10.1186/s12864-015-1419-2]
Pandey P.K., Dhotre D.P., Dharne M.S., Khadse A.N., Hiremath U.I., Chaudhari R.D., Patole M.S., Shouche Y.S. (2007). Evaluation of mitochondrial 12S rRNA gene in the identification of Panterapardusfusca (Meyer, 1794) from field-collected scat samples in Western Ghats, Maharashtra, India. Current Science. 92: 1129-1133.
Pang Z., Zhou G., Ewald J., Chang L., Haçarız O., Basu N., Xia J. (2022). Using MetaboAnalyst 5.0 for LC–HRMS spectra processing, multi-omics integration, and covariate adjustment of global metabolomics data. Nature Protocols. 17: 1735-1761. [DOI: 10.1038/s41596-022-00715-4]
Pillay S., De Vos M., Derendinger B., Streicher E.M., Dolby T., Scott L.A., Steinhobel A.D., Warren R.M., Theron G. (2022). Non-actionable results, accuracy, and effect of first- and second-line line probe assays for diagnosing drug-resistant tuberculosis, including on smear-negative specimens, in a high-volume laboratory. Clinical Infectious Diseases. 76: e920–e929. [DOI: 10.1093/cid/ciac556]
Prakrongrak N., Boonsoong B., Monthatong M. (2023). Genetic diversity and phylogenetic analysis of mayfly Caenis (Insecta: Ephemeroptera) using Cytochrome C Oxidase I (COI) and 12s rRNA genes from Thailand. Biodiversitas Journal of Biological Diversity. 24: 1942-1952. [DOI: 10.13057/biodiv/ d240407]
Rahman M.M., Ahmad Z., Mustafa S. (2023). DNA-based platform for halal authentication and combat food threat. Journal of Halal Industry and Services. [DOI: 10.36877/jhis.a0000407]
Reuter J., Spacek D.V., Snyder M. (2015).High-throughput sequencing technologies. Molecular Cell. 58: 586–597. [DOI: 10.1016/j.molcel.2015.05.004]
Rhoads A., Au K.F. (2015). PacBio sequencing and its applications. Genomics, Proteomics and Bioinformatics. 13: 278–289. [DOI: 10.1016/j.gpb. 2015.08.002]
Ripp F., Krombholz C.F., Liu Y., Weber M., Schäfer A., Schmidt B., Köppel R., Hankeln T. (2014). All-Food-Seq (AFS): a quantifiable screen for species in biological samples by deep DNA sequencing. BMC Genomics. 15: 639. [DOI: 10.1186/1471-2164-15-639]
Rohman A., Orbayinah S., Hermawan A., Sudjadi S., Windarsih A., Handayani S. (2022). The development of real-time polymerase chain reaction for identification of beef meatball. Applied Food Research. 2: 100148. [DOI: 10.1016/j.afres.2022.100148]
Roy D., Akhter S., Sarker A., Hossain M., Lyzu C., Mohanta L., Islam D., Khan M. (2022). Tracing the pig and cattle origin in processed food and feed products targeting mitochondrial 12S rRNA gene. Journal of Food Quality and Hazards Control. 9: 163-171. [DOI: 10.18502/jfqhc.8.4.8256]
Sahilah A.M., Norhayati Y., Norrakiah A.S., Aminah A., Wan Aida W.M. (2011). Halal authentication of raw meats using PCR amplification of mitochondrial DNA. International Food Research Journal. 18: 1489-1491.
Sajali N., Wong S.C., Bakar S.A., Mokhtar N.F.K., Manaf Y.N.A., Yuswan M.H., Desa M.N.M. (2020). Analytical approaches of meat authentication in food. International Journal of Food Science and Technology. 56: 1535–1543. [DOI: 10.1111/ ijfs.14797]
Sanjaya U.H., Arabella R. (2023). Legal protection of consumer data on e-commerce platforms with cash on delivery (COD) systems. KnE Social Sciences. 8: 20-32. [DOI: 10.18502/kss.v8i1.10869]
Satam H., Joshi K., Mangrolia U., Waghoo S., Zaidi G., Rawool S., Thakare R.P., Banday S., Mishra A., Das G., Malonia S.K. (2023). Next-generation sequencing technology: current trends and advancements. Biology.12: 997. [DOI: 10.3390/biology12070997]
Schloss P.D., Gevers D., Westcott S.L. (2011). Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLOS ONE. 6: e27310. [DOI: 10.1371/journal.pone.0027310]
Sedlazeck F.J., Rescheneder P., Smolka M., Han F., Nattestad M., Von Haeseler A., Schatz M.C. (2018).Accurate detection of complex structural variations using single-molecule sequencing. Nature Methods. 15: 461–468. [DOI: 10.1038/s41592-018-0001-7]
Segata N., Waldron L., Ballarini A., Narasimhan V., Jousson O., Huttenhower C. (2012). Metagenomic microbial community profiling using unique clade-specific marker genes. Nature Methods. 9: 811–814. [DOI: 10.1038/nmeth.2066]
Shaharuddin F.Z., Norman A.A., Hamid S., Zakaria Z. (2025). Halal food industry in Malaysia: issues and challenges. Food Research. 9: 33–42. [DOI: 10.26656/fr.2017.9(s2).072]
Shen W., Song Z., Zhong X., Huang M., Shen D., Gao P., Qian X., Wang M., He X., Wang T., Li S., Song X. (2022). Sangerbox: a comprehensive, interaction‐friendly clinical bioinformatics analysis platform. iMeta. 1: 15. [DOI: 10.1002/imt2.15]
Shirazi S., Meyer R.S., Shapiro B. (2021). Revisiting the effect of PCR replication and sequencing depth on biodiversity metrics in environmental DNA metabarcoding. Ecology and Evolution. 11: 15766–15779. [DOI: 10.1002/ece3.8239]
Slatko B.E., Gardner A.F., Ausubel F.M. (2018).Overview of next-generation sequencing technologies. Current Protocols in Molecular Biology. 122: e59. [DOI: 10.1002/cpmb.59]
Sun K., Liu Y., Zhou X., Yin C., Zhang P., Yang Q., Mao L., Shentu X., Yu X. (2022). Nanopore sequencing technology and its application in plant virus diagnostics. Frontiers in Microbiology. 13: 939666. [DOI: 10.3389/fmicb.2022.939666]
Taber K.S. (2017). The use of Cronbach’s Alpha when developing and reporting research instruments in science education. Research in Science Education. 48: 1273–1296. [DOI: 10.1007/s11165-016-9602-2]
Talib M.S.A., Zulfakar M.H. (2023). Sustainable halal food supply chain management in a small rentier halal market. Arab Gulf Journal of Scientific Research. 41: 61-72. [DOI: 10.1108/agjsr-11-2022-0251]
Teng J.L.L., Yeung M.L., Chan E., Jia L., Lin C.H., Huang Y., Tse H., Wong S.S.Y., Sham P.C., Lau S.K.P., Woo P.C.Y. (2017). PacBio but not Illumina technology can achieve fast, accurate and
complete closure of the high GC, complex Burkholderiapseudomallei two-chromosome genome. Frontiers in Microbiology. 8: 1448. [DOI: 10.3389/fmicb.2017.01448]
Thomas G.W.C., Hahn M.W. (2019). Referee: reference assembly quality scores. Genome Biology and Evolution. 11: 1483–1486. [DOI: 10.1093/ gbe/evz088]
Tyler A.D., Mataseje L., Urfano C., Schmidt L., Antonation K., Mulvey M.R., Corbett C.R. (2018). Evaluation of Oxford Nanopore’sMinION sequencing device for microbial whole genome sequencing applications. Scientific Reports. 8: 10931. [DOI: 10.1038/s41598-018-29334-5]
Udaondo Z., Sittikankaew K., Uengwetwanit T., Wongsurawat T., Sonthirod C., Jenjaroenpun P., Pootakham W., Karoonuthaisiri N., Nookaew I. (2021). Comparative analysis of PacBio and Oxford Nanopore sequencing technologies for transcriptomic landscape identification of Penaeusmonodon. Life. 11: 862. [DOI: 10.3390/life11080862]
Uliano-Silva M., Ferreira J.G.R.N., Krasheninnikova K., Blaxter M., Mieszkowska N., Hall N., Holland P., Durbin R., Richards T., Kersey P., Hollingsworth P., Wilson W., Twyford A., Gaya E., Lawniczak M., Lewis O., Broad G., Martin F., Hart M., McCarthy S.A. (2023). MitoHiFi: a python pipeline for mitochondrial genome assembly from PacBio high fidelity reads. BMC Bioinformatics. 24: 288. [DOI: 10.1186/s12859-023-05385-y]
Van Der Spiegel M., Van Der Fels-Klerx H., Sterrenburg P., Van Ruth S.M., Scholtens-Toma I., Kok E. (2012). Halal assurance in food supply chains: verification of halal certificates using audits and laboratory analysis. Trends in Food Science and Technology. 27: 109–119. [DOI: 10.1016/j.tifs.2012.04.005]
Venkatas J., Adeleke M.A. (2019). Emerging threat of Eimeria operational taxonomic units (OTUs) on poultry production. Parasitology. 146: 1615–1619. [DOI: 10.1017/s0031182019001100]
Wang H., Chen Y., Wang L., Liu Q., Yang S., Wang C. (2023). Advancing herbal medicine: enhancing product quality and safety through robust quality control practices. Frontiers in Pharmacology. 14: 1265178. [DOI: 10.3389/fphar.2023.1263813]
Wang X., Cai Y., Sun Y., Knight R., Mai V. (2011). Secondary structure information does not improve OTU assignment for partial 16S rRNA sequences. The ISME Journal. 6: 1277–1280. [DOI: 10.1038/ismej.2011.187]
Wasswa F.B., Kassaza K., Nielsen K., Bazira J. (2022). MinION whole-genome sequencing in resource-limited settings: challenges and opportunities. Current Clinical Microbiology Reports. 9: 52–59. [DOI: 10.1007/s40588-022-00183-1]
Wenger A., Peluso P., Rowell W., Chang P.C., Hall R., Concepcion G., Hunkapiller M. (2019). Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nature Biotechnology. 37: 1155–1162. [DOI: 10.1038/s41587-019-0217-9]
Wick R.R., Judd L.M., Holt K.E. (2023). Assembling the perfect bacterial genome using Oxford Nanopore and Illumina sequencing. PLOS Computational Biology. 19: e1010905. [DOI: 10.1371/journal.pcbi.1010905]
Woltyńska A., Gawor J., Olech M.A., Górniak D., Grzesiak J. (2023). Bacterial communities of Antarctic lichens explored by gDNA and cDNA 16S rRNA gene amplicon sequencing. FEMS Microbiology Ecology. 99: fiad015. [DOI: 10.1093/femsec/fiad015]
Woo C., Kumari P., Eo K., Lee W., Kimura J., Yamamoto N. (2023). Combining vertebrate mitochondrial 12S rRNA gene sequencing and shotgun metagenomic sequencing to investigate the diet of the leopard cat (Prionailurusbengalensis) in Korea. PLOS ONE. 18: e0278941. [DOI: 10.1371/journal.pone.0278941]
Wood D.E., Lu J., Langmead B. (2019). Improved metagenomic analysis with Kraken 2. Genome Biology. 20: 257. [DOI: 10.1186/s13059-019-1891-0]
Yakin A.U. (2021). Halal certification, standards, and their ramifications in Belgium. Islamic Studies Journal. 27: 45-59. [DOI: 10.1163/9789004459236_ 008]
Yang Y., Xie B., Yan J. (2014). Application of next-generation sequencing technology in forensic science. Genomics, Proteomics and Bioinformatics. 12: 190–197. [DOI: 10.1016/j.gpb.2014.09.001]
Yang L., Tan Z., Wang D., Xue L. Guan M., Huang T., Li R. (2014). Species identification through mitochondrial rRNA genetic analysis. Scientific Reports. 4: 4089. [DOI: 10.1038/srep04089]
Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., Madden T.L. (2012). Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics.13: 134. [DOI: 10.1186/ 1471-2105-13-134]
Zhang Y., Qu Q., Mingzhen R., Nana Z., Zhao Y., Tao F. (2020). Simultaneous identification of animal-derived components in meats using high-throughput sequencing in combination with a custom-built mitochondrial genome database. Scientific Reports.10: 8965. [DOI: 10.1038/s41598-020-65724-4]
Zhou W., Liu X., Lv M., Shi Y., Zhang L. (2023). The recognition mode between hsRBFA and mitoribosome 12S rRNA during mitoribosomal biogenesis. Nucleic Acids Research. 51: 1353–1363. [DOI: 10.1093/ nar/gkac1234]
Zou A., Phan L., Chen S., Campbell J., Guo P., Ren R., Hendrycks D. (2023). Representation engineering: a top-down approach to AI transparency. arXiv. [DOI: 10.48550/arXiv.2301.01912]
[*]Corresponding author (J. Jani)
E-mail: jaeyres@ums.edu.my
ORCID ID: https://orcid.org/0000-0001-8956-0538
E-mail: jaeyres@ums.edu.my
ORCID ID: https://orcid.org/0000-0001-8956-0538
Type of Study: Review article |
Subject:
Special
Received: 25/04/20 | Accepted: 25/11/25 | Published: 25/12/21
Received: 25/04/20 | Accepted: 25/11/25 | Published: 25/12/21
References
1. Absmeier E., Chandrasekaran V., O'Reilly F.J., Stowell J.a.W., Rappsilber J., Passmore L.A. (2023). Specific recognition and ubiquitination of translating ribosomes by mammalian CCR4-NOT. Nature Structural and Molecular Biology. 30: 1314-1322. [DOI: 10.1038/s41594-023-01075-8] [DOI:10.1038/s41594-023-01075-8] [PMID] [PMCID]
2. Ahsan M.U., Liu Q., Perdomo J.E., Fang L., Wang K. (2023). A survey of algorithms for the detection of genomic structural variants from long-read sequencing data. Nature Methods. 20: 1143-1158. [DOI: 10.1038/s41592-023-01932-w] [DOI:10.1038/s41592-023-01932-w] [PMID] [PMCID]
3. Aini S.R., Rohman A., Mulyanto M., Erwanto Y., Ansar A., Handayani B.R., Irnawati I. (2023). The metabolomics approach used for halal authentication analysis of food and pharmaceutical products: a review. Food Research. 7: 180-187. [DOI:10.26656/fr.2017.7(3).986]
4. Aksamentov I., Roemer C., Hodcroft E.B., Neher R.A. (2021). Nextclade: clade assignment, mutation calling and quality control for viral genomes. Journal of Open Source Software. 6: 3773. [DOI: 10.21105/joss.03773] [DOI:10.21105/joss.03773]
5. Aksin N., Aini F.N.Q. (2023). Halal Ṭayyib syaria’ in understanding the needs of consumer protection. Jurnal Meta-Yuridis. 6: 1-13. [DOI: 10.26877/m-y.v6i2.15560] [DOI:10.26877/m-y.v6i2.15560]
6. Algan F.M. (2023). Global standardization for a sustainable future. Current Research in Social Sciences. 9: 30-40. [DOI: 10.30613/ curesosc.1173926] [DOI:10.30613/curesosc.1173926]
7. Allam Z., Bibri S.E., Chabaud D., Moreno C. (2022). The theoretical, practical, and technological foundations of the 15-minute city model: proximity and its environmental, social, and economic benefits for sustainability. Energies. 15: 6042. [DOI: 10.3390/ en15166042] [DOI:10.3390/en15166042]
8. Amarasinghe S.L., Su S., Dong X., Zappia L., Ritchie M.E., Gouil Q. (2020). Opportunities and challenges in long-read sequencing data analysis. Genome Biology. 21: 30. [DOI: 10.1186/s13059-020-1935-5] [DOI:10.1186/s13059-020-1935-5] [PMID] [PMCID]
9. Amer M. (2024). Halal standards' implementation in Palestinian food sector: its drivers and impact on performance. Arab Gulf Journal of Scientific Research. 42: 2-29. [DOI: 10.1108/agjsr-09-2022-0168] [DOI:10.1108/AGJSR-09-2022-0168]
10. Aziz N., Bakry N., Habibi Mz M., Siddiq Armia M. (2023). The paradigm of modern food products and its relevance with the concept of food in the Quran. Heliyon. 9: e21358. [DOI: 10.1016/j. heliyon.2023.e21358] [DOI:10.1016/j.heliyon.2023.e21358] [PMID] [PMCID]
11. Bannor R.K., Arthur K.E., Oppong D., Oppong-Kyeremeh H. (2023). A comprehensive systematic review and bibliometric analysis of food fraud from a global perspective. Journal of Agriculture and Food Research. 14: 100686. [DOI: 10.1016/j.jafr. 2023.100686] [DOI:10.1016/j.jafr.2023.100686]
12. Bianconi I., Aschbacher R., Pagani E. (2023). Current uses and future perspectives of genomic technologies in clinical microbiology. Antibiotics. 12: 1580. [DOI: 10.3390/antibiotics12111580] [DOI:10.3390/antibiotics12111580] [PMID] [PMCID]
13. Bohmann K., Elbrecht V., Carøe C., Bista I., Leese F., Bunce M., Yu D.W., Seymour M., Dumbrell A.J., Creer S. (2022). Strategies for sample labelling and library preparation in DNA metabarcoding studies. Molecular Ecology Resources. 22: 1231-1246. [DOI: 10.1111/1755-0998.13512] [DOI:10.1111/1755-0998.13512] [PMID] [PMCID]
14. Bolger A.M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30: 2114-2120. [DOI: 10.1093/bioinformatics/btu170] [DOI:10.1093/bioinformatics/btu170] [PMID] [PMCID]
15. Borodinov A., Manoilov V., Zarutsky I., Petrov A., Kurochkin V., Saraev A. (2022). Machinelearning in base-calling for next-generation sequencing methods. Informatics and Automation. 21: 572-603. [DOI: 10.15622/ia.21.3.5] [DOI:10.15622/ia.21.3.5]
16. Bosona T., Gebresenbet G. (2023). The role of blockchain technology in promoting traceability systems in agri-food production and supply chains. Sensors. 23: 5342. [DOI: 10.3390/s23115342] [DOI:10.3390/s23115342] [PMID] [PMCID]
17. Boyrusbianto F.T., Utama D.T., Khikmawati I., Fadhila A., Volkandari S.D., Abdurrahman Z.H., Pramono A., Cahyadi M. (2023). A simplex and multiplex PCR assay for simultaneous detection of beef, pork, and chicken meat in sausages based on mitochondrial DNA Cytochrome oxidase sub-unit I. Food Research. 7: 188-193. [DOI: 10.26656/fr.2017.7(3).084] [DOI:10.26656/fr.2017.7(3).084]
18. Cahyadi M., Wibowo T., Pramono A., Abdurrahman Z.H. (2020). A novel multiplex-PCR assay to detect three non-halal meats contained in meatballs using mitochondrial 12S rRNA gene. Food Science of Animal Resources. 40: 628-635. [DOI: 10.5851/kosfa.2020.e40] [DOI:10.5851/kosfa.2020.e40] [PMID] [PMCID]
19. Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nature Methods. 13: 581-583. [DOI: 10.1038/ nmeth.3869] [DOI:10.1038/nmeth.3869] [PMID] [PMCID]
20. Cantalapiedra C.P., Hernández-Plaza A., Letunic I., Bork P., Huerta-Cepas J. (2021). eggNOG-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Molecular Biology and Evolution. 38: 5825-5829. [DOI: 10.1093/molbev/msab293] [DOI:10.1093/molbev/msab293] [PMID] [PMCID]
21. Cao M.D., Ganesamoorthy D., Cooper M.A., Coin L.J.M. (2015). Realtime analysis and visualization of MinION sequencing data with npReader. Bioinformatics. 32: 764-766. [DOI: 10.1093/bioinformatics/btv658] [DOI:10.1093/bioinformatics/btv658] [PMID]
22. Chan A.H.E., Saralamba N., Saralamba S., Ruangsittichai J., Thaenkham U. (2022). The potential use of mitochondrial ribosomal genes (12S and 16S) in DNA barcoding and phylogenetic analysis of trematodes. BMC Genomics. 23: 104. [DOI: 10.1186/s12864-022-08302-4] [DOI:10.1186/s12864-022-08302-4] [PMID] [PMCID]
23. Chang J.J.M., Ip Y.C.A., Bauman A.G., Huang D. (2020). MINION-in-ARMS: nanopore sequencing to expedite barcoding of specimen-rich macrofaunal samples from autonomous reef monitoring structures. Frontiers in Marine Science. 7: 448. [DOI: 10.3389/fmars.2020. 00448] [DOI:10.3389/fmars.2020.00448]
24. Chen C., Wu Y., Li J., Wang X., Zeng Z., Xu J., Liu Y., Feng J., Chen H., He Y., Xia R. (2023). TBtools-II: a "one for all, all for one" bioinformatics platform for biological big-data mining. Molecular Plant. 16: 1733-1742. [DOI: 10.1016/j.molp.2023.09.010] [DOI:10.1016/j.molp.2023.09.010] [PMID]
25. Chen S., Zhou Y., Chen Y., Gu J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34: i884-i890. [DOI: 10.1093/bioinformatics/bty560] [DOI:10.1093/bioinformatics/bty560] [PMID] [PMCID]
26. Cobbe J., Veale M., Singh J. (2023). Understanding accountability in algorithmic supply chains. Proceedings of the 2023 ACM Conference on Fairness, Accountability, and Transparency. 1186-1197. [DOI: 10.1145/3593013.3594073] [DOI:10.1145/3593013.3594073]
27. Daniel A.I., Fadaka A.O., Gokul A., Bakare O.O., Aina O., Fisher S., Burt A.F., Mavumengwana V., Keyster M., Klein A. (2022). Biofertilizer: the future of food security and food safety. Microorganisms. 10: 1220. [DOI: 10.3390/microorganisms10061220] [DOI:10.3390/microorganisms10061220] [PMID] [PMCID]
28. De Coster W., Weissensteiner M.H., Sedlazeck F.J. (2021). Towards population-scale long-read sequencing. Nature Reviews Genetics. 22: 572-587. [DOI: 10.1038/s41576-021-00367-3] [DOI:10.1038/s41576-021-00367-3] [PMID] [PMCID]
29. Delahoy M.J., Shah H.J., Weller D.L., Ray L.C., Smith K., McGuire S., Trevejo R.T., Walter E.S., Wymore K., Rissman T., McMillian M., Lathrop S., et al. (2023). Preliminary incidence and trends of infections caused by pathogens transmitted commonly through food - foodborne diseases active surveillance network, 10 U.S. sites, 2022. Morbidity and Mortality Weekly Report. 72: 701-706. [DOI: 10.15585/ mmwr.mm7226a1] [DOI:10.15585/mmwr.mm7226a1] [PMID] [PMCID]
30. Dubois B., Debode F., Hautier L., Hulin J., Martin G.S., Delvaux A., Janssen E., Mingeot D. (2022). A detailed workflow to develop QIIME2-formatted reference databases for taxonomic analysis of DNA metabarcoding data. BMC Genomic Data. 23: 53. [DOI: 10.1186/s12863-022-01045-w] [DOI:10.1186/s12863-022-01067-5] [PMID] [PMCID]
31. Dunham J.P., Friesen M.L. (2013). A cost-effective method for high-throughput construction of Illumina sequencing libraries. Cold Spring Harbor Protocols. 2013: 820-834. [DOI: 10.1101/pdb.prot074187] [DOI:10.1101/pdb.prot074187] [PMID] [PMCID]
32. El-Nabi S.E.H., Hussein D., Khallafa A.G. (2021). Molecular detection of food fraud targeting mitochondrial 12S rRNA gene sequencing. Journal of Bioscience and Applied Research. 7: 17-22. [DOI: 10.21608/jbaar.2021.167091] [DOI:10.21608/jbaar.2021.167091]
33. Ezeaku H.C., Asongu S.A., Nnanna J. (2021). Volatility of international commodity prices in times of COVID-19: effects of oil supply and global demand shocks. The Extractive Industries and Society. 8: 257-270. [DOI: 10.1016/j.exis.2020.12.013] [DOI:10.1016/j.exis.2020.12.013]
34. Ferreira T., Rodriguez S. (2024). Mitochondrial DNA: inherent complexities relevant to genetic analyses. Genes. 15: 617. [DOI: 10.3390/ genes15050617] [DOI:10.3390/genes15050617] [PMID] [PMCID]
35. Fosso B., Santamaria M., Marzano M., Alonso-Alemany D., Valiente G., Donvito G., Monaco A., Notarangelo P., Pesole G. (2015). BioMaS: a modular pipeline for bioinformatic analysis of metagenomic amplicons. BMC Bioinformatics. 16: 203. [DOI: 10.1186/s12859-015-0595-z] [DOI:10.1186/s12859-015-0595-z] [PMID] [PMCID]
36. Fu L., Niu B., Zhu Z., Wu S., Li W. (2012). CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 28: 3150-3152. [DOI: 10.1093/bioinformatics/bts565] [DOI:10.1093/bioinformatics/bts565] [PMID] [PMCID]
37. Galal-Khallaf A. (2021). Multiplex PCR and 12S rRNA gene sequencing for detection of meat adulteration: a case study in the Egyptian markets. Gene. 764: 145062. [DOI: 10.1016/j.gene.2020.145062] [DOI:10.1016/j.gene.2020.145062] [PMID]
38. Gallardo A., Choy C., Juneja J., Bozkir E., Cobb C., Bauer L., Cranor L. (2023). Speculative privacy concerns about AR glasses data collection. Proceedings on Privacy Enhancing Technologies. 2023: 416-435. [DOI: 10.56553/ popets-2023-0117] [DOI:10.56553/popets-2023-0117]
39. Garinet S., Laurent-Puig P., Blons H., Oudart J. (2018). Current and future molecular testing in NSCLC: what can we expect from new sequencing technologies? Journal of Clinical Medicine. 7: 144. [DOI: 10.3390/jcm7060144] [DOI:10.3390/jcm7060144] [PMID] [PMCID]
40. Gupta N., Verma V. (2019). Next-generation sequencing and its application: empowering in public health beyond reality. In: Arora P. (Editor). Microbial technology for the welfare of society. Springer, Singapore. pp: 313-341. [DOI: 10.1007/978-981-13-8844-6_15] [DOI:10.1007/978-981-13-8844-6_15] [PMCID]
41. Hassan N., Ahmad T., Zain N.M., Ashaari A. (2020). A novel chemometrics method for halal authentication of gelatin in food products. SainsMalaysiana. 49: 2083-2089. [DOI: 10.17576/jsm-2020-4907-08] [DOI:10.17576/jsm-2020-4907-08]
42. Hayes B., Nguyen L.T., Forutan M., Engle B.N., Lamb H.J., Copley J.P., Randhawa I.A.S., Ross E.M. (2021). An epigenetic aging clock for cattle using portable sequencing technology. Frontiers in Genetics. 18: 760450. [DOI: 10.3389/fgene. 2021.760450] [DOI:10.3389/fgene.2021.760450] [PMID] [PMCID]
43. Haynes E., Jiménez E., Pardo M.Á., Helyar S.J. (2019). The future of NGS (Next Generation Sequencing) analysis in testing food authenticity. Food Control. 101: 134-143. [DOI: 10.1016/j.foodcont. 2019.02. 010] [DOI:10.1016/j.foodcont.2019.02.010]
44. Hernández V.P., Guzmán M.H. (2023). Microorganisms identification in environmental samples: bioinformatic analysis of 16S rRNA gene with QIIME2. Ciencia Latina Revista Científica Multidisciplinar. 7: 8074-8102. [DOI: 10.37811/cl_rcm.v7i5.8382] [DOI:10.37811/cl_rcm.v7i5.8382]
45. Hidayat H.H., Wijayanti N., Situmorang S.C.D.U.B. (2023). Materials clustering of Soto Sokaraja as an effort to accelerate halal certification. IOP Conference Series: Earth and Environmental Science. 1177: 012036. [DOI: 10.1088/1755-1315/1177/ 1/012036] [DOI:10.1088/1755-1315/1177/1/012036]
46. Hook P.W., Timp W. (2023). Beyond assembly: the increasing flexibility of single-molecule sequencing technology. Nature Reviews Genetics. 24: 627-641. [DOI: 10.1038/s41576-023-00600-1] [DOI:10.1038/s41576-023-00600-1] [PMID] [PMCID]
47. Jani N.J., Chin N.K.L., Mustapha N.Z.A. (2023). Whole genome sequence analysis of a Mycobacterium iranicum, a newly identified non-tuberculosis mycobacteria, strain SBH312 isolated in Sabah, Malaysia. Borneo Journal of Medical Sciences. 17: 52-56. [DOI: 10.51200/bjms.v17i1.4291] [DOI:10.51200/bjms.v17i1.4291]
48. Jaswir I., Mirghani M.E.S., Salleh H.M., Ramli N., Octavianti F., Hendri R. (2016). An overview of the current analytical methods for halal testing. Current issues in halal food analysis. Springer, Singapore. pp: 291-300. [DOI: 10.1007/978-981-10-1452-9_27] [DOI:10.1007/978-981-10-1452-9_27]
49. Jiang L. (2023). Environmental benefits of green buildings with BIM technology. Ecological Chemistry and Engineering. 30: 191-199. [DOI: 10.1515/eces-2023-0003] [DOI:10.2478/eces-2023-0019]
50. Jovic D., Liang X., Zeng H., Lin L., Xu F., Luo Y. (2022). Single‐cell RNA sequencing technologies and applications: a brief overview. Clinical and Translational Medicine. 12: 21-35. [DOI: 10.1002/ ctm2.983] [DOI:10.1002/ctm2.694] [PMID] [PMCID]
51. Karst S.M., Ziels R.M., Kirkegaard R., Sørensen E.A., McDonald D., Zhu Q., Knight R., Albertsen M. (2021). High-accuracy long-read amplicon sequences using unique molecular identifiers with Nanopore or PacBio sequencing. Nature Methods. 18: 165-169. [DOI: 10.1038/s41592-020-01041-y] [DOI:10.1038/s41592-020-01041-y] [PMID]
52. Katoh K., Rozewicki J., Yamada K.D. (2017). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics. 20: 1160-1166. [DOI: 10.1093/bib/bbx108] [DOI:10.1093/bib/bbx108] [PMID] [PMCID]
53. Kaveh S., Hashemi S.M.B., Abedi E., Amiri M.J., Conte F.L. (2023). Bio-preservation of meat and fermented meat products by lactic acid bacteria strains and their antibacterial metabolites. Sustainability. 15: 10154. [DOI: 10.3390/su151310154] [DOI:10.3390/su151310154]
54. Kayama K., Kanno M., Chisaki N., Tanaka M., Yao R., Hanazono K., Camer G.A., Endoh D. (2021). Prediction of PCR amplification from primer and template sequences using recurrent neural network. Scientific Reports. 11: 7493. [DOI: 10.1038/s41598-021-86357-1] [DOI:10.1038/s41598-021-86357-1] [PMID] [PMCID]
55. King J., Harder T., Beer M., Pohlmann A. (2020). Rapid multiplex MinION nanopore sequencing workflow for Influenza A viruses. BMC Infectious Diseases. 20: 648. [DOI: 10.1186/s12879-020-05367-y] [DOI:10.1186/s12879-020-05367-y] [PMID] [PMCID]
56. Kozich J.J., Westcott S.L., Baxter N.T., Highlander S.K., Schloss P.D. (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MISEQ Illumina sequencing platform. Applied and Environmental Microbiology. 79: 5112-5120. [DOI: 10.1128/aem. 01043-13] [DOI:10.1128/AEM.01043-13] [PMID] [PMCID]
57. Krisna R., Yusuf M. (2023). Halal ecosystem improvement study reviewed of halal product regulations. International Journal of Research and Review. 10: 339-353. [DOI: 10.52403/ijrr.20230243] [DOI:10.52403/ijrr.20230243]
58. Kumari P., Dong K., Eo K., Lee W., Kimura J., Yamamoto N. (2019). DNA metabarcoding-based diet survey for the Eurasian otter (Lutralutra): Development of a Eurasian otter-specific blocking oligonucleotide for 12S rRNA gene sequencing for vertebrates. PLOS ONE. 14: e0226253. [DOI: 10.1371/journal.pone.0222966] [DOI:10.1371/journal.pone.0226253] [PMID] [PMCID]
59. Liu H., Nie J., Liu Y., Wadood S.A., Rogers K.M., Yuan Y., Gan R. (2023). A review of recent compound-specific isotope analysis studies applied to food authentication. Food Chemistry. 415: 135791. [DOI: 10.1016/j.foodchem.2023.135791] [DOI:10.1016/j.foodchem.2023.135791] [PMID]
60. Liu J., Seetharam A.S., Chougule K., Ou S., Swentowsky K.W., Gent J.I., Llaca V., Woodhouse M.R., Manchanda N., Presting G.G., Kudrna D.A., Alabady M., Hirsch C.N., Fengler K.A., Ware D., Michael T.P., Hufford M.B., Dawe R.K. (2020). Gapless assembly of maize chromosomes using long-read technologies. Genome Biology. 21: 121. [DOI: 10.1186/s13059-020-02029-9] [DOI:10.1186/s13059-020-02029-9] [PMID] [PMCID]
61. Liu X., Liu Z., Cheng Y., Wu H., Shen W., Liu Y., Feng Q., Yang J. (2021). Application of next-generation sequencing technology based on single gene locus in species identification of mixed meat products. Journal of Food Quality. 2021: 4512536. [DOI: 10. 1155/2021/4512536] [DOI:10.1155/2021/4512536]
62. Lu H., Giordano F., Ning Z. (2016). Oxford NanoporeMinION sequencing and genome assembly. Genomics, Proteomics and Bioinformatics. 14: 265-279. [DOI: 10.1016/j.gpb.2016.05.004] [DOI:10.1016/j.gpb.2016.05.004] [PMID] [PMCID]
63. Lubis H., Mohd-Naim N.F., Alizul N.N., Ahmed M.U. (2016). From market to food plate: current trusted technology and innovations in halal food analysis. Trends in Food Science and Technology. 58: 55-68. [DOI: 10.1016/j.tifs.2016.10.024] [DOI:10.1016/j.tifs.2016.10.024]
64. Lücking R., Aime M.C., Robbertse B., Miller A., Ariyawansa H., Aoki T., Papp V., Robert V., Schoch C. (2020). Unambiguous identification of fungi: where do we stand and how accurate and precise is fungal DNA barcoding? IMA Fungus. 11: 14. [DOI: 10.1186/s43008-020-00033-9] [DOI:10.1186/s43008-020-00033-z] [PMID] [PMCID]
65. Luo J., Yan D., Zhang D., Han Y., Dong X., Yang Y., Deng K., Xiao X. (2011). Application of 12S rRNA gene for the identification of animal-derived drugs. Journal of Pharmacy and Pharmaceutical Sciences. 14: 358-367. [DOI: 10.18433/j3n017] [DOI:10.18433/J3N017] [PMID]
66. Ma X., Shao Y., Tian L., Flasch D.A., Mulder H.L., Edmonson M.N., Liu Y., Chen X., Newman S., Nakitandwe J., Li Y., Li B., Shen S., Wang Z., Shurtleff S., Robison L.L., Levy S., Easton J., Zhang J. (2019). Analysis of error profiles in deep next-generation sequencing data. Genome Biology. 20: 50. [DOI: 10.1186/s13059-019-1659-6] [DOI:10.1186/s13059-019-1659-6] [PMID] [PMCID]
67. Malaysia's Halal Food Opportunities (MIDA). (2022). Malaysia's halal food opportunities - carving out the lion's share in a USD3 trillion global market. URL: https://www.mida.gov.my/malaysias-halal-food-opportunitiescarving-out-the-lions-share-in-a-usd3-trillion-global-market/. Accessed on 5 January 2025.
68. Mathew J.L. (2023). Efficiency, economy, and excellence: experimental exploration of evidence-based guideline development in resource-constrained settings. Indian Journal of Pediatrics. 90: 700-707. [DOI: 10.1007/s12098-023-03785-1] [DOI:10.1007/s12098-023-04575-z] [PMID] [PMCID]
69. Matsunaga T., Chikuni K., Tanabe R., Muroya S., Shibata K., Yamada J., Shinmura Y. (1999). A quick and simple method for the identification of meat species and meat products by PCR assay. Meat Science. 51: 143-148. [DOI: 10.1016/s0309-1740(98)00112-0] [DOI:10.1016/S0309-1740(98)00112-0] [PMID]
70. Menzel P., Ng K.L., Krogh A. (2016). Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nature Communications. 7: 11257. [DOI: 10.1038/ncomms11257] [DOI:10.1038/ncomms11257] [PMID] [PMCID]
71. Meyer M., Kircher M. (2010). Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harbor Protocols. 2010: pdb.prot5448. [DOI: 10.1101/pdb.prot5448] [DOI:10.1101/pdb.prot5448] [PMID]
72. Mitchell A.L., Almeida A., Beracochea M., Boland M., Burgin J., Cochrane G., Crusoe M.R., Kale V., Potter S.C., Richardson L.J., Sakharova E., Scheremetjew M., et al. (2020). MGnify: the microbiome analysis resource in 2020. Nucleic Acids Research. 48: D570-D578. [DOI: 10.1093/nar/gkz1035] [DOI:10.1093/nar/gkz1035] [PMID] [PMCID]
73. Moorthie S., Mattocks C., Wright C.F. (2011). Review of massively parallel DNA sequencing technologies. The HUGO Journal. 5: 1-12. [DOI: 10.1007/s11568-011-9156-3] [DOI:10.1007/s11568-011-9156-3] [PMID] [PMCID]
74. Montiel-Sosa J.F., Ruiz-Pesini E., Montoya J., Roncalés P., López-Pérez M.J., Pérez-Martos A. (2000). Direct and highly species-specific detection of pork meat and fat in meat products by PCR amplification of mitochondrial DNA. Journal of Agricultural and Food Chemistry. 48: 2829-2832. [DOI: 10.1021/jf9907438] [DOI:10.1021/jf9907438] [PMID]
75. Monzel A.S., Enríquez J.A., Picard M. (2023). Multifaceted mitochondria: moving mitochondrial science beyond function and dysfunction. Nature Metabolism. 5: 546-562. [DOI: 10.1038/s42255-023-00783-1] [DOI:10.1038/s42255-023-00783-1] [PMID] [PMCID]
76. Muflihah, Hardianto A., Kusumaningtyas P., Prabowo S., Hartati Y.W. (2023). DNA-based detection of pork content in food. Heliyon. 9: e14418. [DOI: 10.1016/j.heliyon.2023.e14418] [DOI:10.1016/j.heliyon.2023.e14418] [PMID] [PMCID]
77. Muralidharan S., Yoo B., Ko H. (2023). Decentralized ME-centric framework-a futuristic architecture for consumer IoT. IEEE Consumer Electronics Magazine. 12: 39-50. [DOI: 10.1109/ MCE. 2023.3152809] [DOI:10.1109/MCE.2022.3151023]
78. Murugaiah C., Noor Z.M., Mastakim M., Bilung L.M., Selamat J., Radu S. (2009). Meat species identification and halal authentication analysis using mitochondrial DNA. Meat Science. 83: 57-61. [DOI: 10.1016/j.meatsci.2009.03.015] [DOI:10.1016/j.meatsci.2009.03.015] [PMID]
79. Nasir S., Zulfakar M.H., Rahmat A. (2021). Determinants of preferred export logistics gateway in Malaysia halal product industry. International Journal of Academic Research in Accounting, Finance and Management Sciences. 11: 60-69. [DOI: 10.6007/IJARAFMS/v11-i4/11823] [DOI:10.6007/IJARAFMS/v11-i1/9823]
80. Neequaye G., Agyekum K., Botchway E. (2017). Market analysis of halal food consumption. International Journal of Food Science. 52: 112-124. [DOI: 10.1002/ijfs.2017.112]
81. Ng P.C., Ruslan N.A.S.A., Chin L.X., Ahmad M., Hanifah S.A., Abdullah Z., Khor S.M. (2021). Recent advances in halal food authentication: challenges and strategies. Journal of Food Science. 87: 8-35. [DOI: 10.1111/1750-3841.15998] [DOI:10.1111/1750-3841.15998] [PMID]
82. Nichols B., Quince C. (2016). Simera: modelling the PCR process to simulate realistic chimera formation. bioRxiv. [DOI: 10.1101/072447] [DOI:10.1101/072447]
83. Nicolae C., Moga L., Bahaciu G., Marin M. (2017). Traceability system structure design for fish and fish products based on supply chain actors' needs. Food Control. 73: 189-200. [DOI: 10.1016/j.foodcont. 2016.10.032]
84. Noakes M.T., Brinkerhoff H., Laszlo A.H., Derrington I.M., Langford K.W., Mount J.W., Bowman J.L., Baker K., Doering K., Tickman B.I., Gundlach J.H. (2019). Increasing the accuracy of nanopore DNA sequencing using a time-varying cross membrane voltage. Nature Biotechnology. 37: 651-656. [DOI: 10.1038/s41587-019-0096-0] [DOI:10.1038/s41587-019-0096-0] [PMID] [PMCID]
85. Noor N.M., Azmi N.A.N., Hanifah N.A., Zamarudin Z. (2023). Navigating the halal food ingredients industry: exploring the present landscape. Halalpshere. 3: 32-43. [DOI: 10.31436/ hs.v3i2.80] [DOI:10.31436/hs.v3i2.80]
86. Nuraini H., Primasari A., Andreas E., Sumantri C. (2012). The use of Cytochrome b gene as a specific marker of the rat meat (Rattusnorvegicus) on meat and meat products. Media Peternakan. 35: 15-20. [DOI: 10.5398/medpet.2012.35.1.15] [DOI:10.5398/medpet.2012.35.1.15]
87. Oehler J.B., Wright H., Stark Z., Mallett A., Schmitz U. (2023). The application of long-read sequencing in clinical settings. Human Genomics. 17: 92. [DOI: 10.1186/s40246-023-00522-3] [DOI:10.1186/s40246-023-00522-3] [PMID] [PMCID]
88. Oktavia L., Marwa T., Yulianita A. (2019). Analysis of factors affecting millennial consumers' demand for halal bread products. MIR (Modernization Innovation Research). 10: 395-407. [DOI: 10.18184/2079-4665.2019.10.3.395-407] [DOI:10.18184/2079-4665.2019.10.3.395-407]
89. Ounit R., Wanamaker S., Close T.J., Lonardi S. (2015). CLARK: fast and accurate classification of metagenomic and genomic sequences using discriminative k-mers. BMC Genomics. 16: 236. [DOI: 10.1186/s12864-015-1419-2] [DOI:10.1186/s12864-015-1419-2] [PMID] [PMCID]
90. Pandey P.K., Dhotre D.P., Dharne M.S., Khadse A.N., Hiremath U.I., Chaudhari R.D., Patole M.S., Shouche Y.S. (2007). Evaluation of mitochondrial 12S rRNA gene in the identification of Panterapardusfusca (Meyer, 1794) from field-collected scat samples in Western Ghats, Maharashtra, India. Current Science. 92: 1129-1133.
91. Pang Z., Zhou G., Ewald J., Chang L., Haçarız O., Basu N., Xia J. (2022). Using MetaboAnalyst 5.0 for LC-HRMS spectra processing, multi-omics integration, and covariate adjustment of global metabolomics data. Nature Protocols. 17: 1735-1761. [DOI: 10.1038/s41596-022-00715-4] [DOI:10.1038/s41596-022-00710-w] [PMID]
92. Pillay S., De Vos M., Derendinger B., Streicher E.M., Dolby T., Scott L.A., Steinhobel A.D., Warren R.M., Theron G. (2022). Non-actionable results, accuracy, and effect of first- and second-line line probe assays for diagnosing drug-resistant tuberculosis, including on smear-negative specimens, in a high-volume laboratory. Clinical Infectious Diseases. 76: e920-e929. [DOI: 10.1093/cid/ciac556] [DOI:10.1093/cid/ciac556] [PMID] [PMCID]
93. Prakrongrak N., Boonsoong B., Monthatong M. (2023). Genetic diversity and phylogenetic analysis of mayfly Caenis (Insecta: Ephemeroptera) using Cytochrome C Oxidase I (COI) and 12s rRNA genes from Thailand. Biodiversitas Journal of Biological Diversity. 24: 1942-1952. [DOI: 10.13057/biodiv/ d240407] [DOI:10.13057/biodiv/d240407]
94. Rahman M.M., Ahmad Z., Mustafa S. (2023). DNA-based platform for halal authentication and combat food threat. Journal of Halal Industry and Services. [DOI: 10.36877/jhis.a0000407] [DOI:10.36877/jhis.a0000407]
95. Reuter J., Spacek D.V., Snyder M. (2015).High-throughput sequencing technologies. Molecular Cell. 58: 586-597. [DOI: 10.1016/j.molcel.2015.05.004] [DOI:10.1016/j.molcel.2015.05.004] [PMID] [PMCID]
96. Rhoads A., Au K.F. (2015). PacBio sequencing and its applications. Genomics, Proteomics and Bioinformatics. 13: 278-289. [DOI: 10.1016/j.gpb. 2015.08.002] [DOI:10.1016/j.gpb.2015.08.002] [PMID] [PMCID]
97. Ripp F., Krombholz C.F., Liu Y., Weber M., Schäfer A., Schmidt B., Köppel R., Hankeln T. (2014). All-Food-Seq (AFS): a quantifiable screen for species in biological samples by deep DNA sequencing. BMC Genomics. 15: 639. [DOI: 10.1186/1471-2164-15-639] [DOI:10.1186/1471-2164-15-639] [PMID] [PMCID]
98. Rohman A., Orbayinah S., Hermawan A., Sudjadi S., Windarsih A., Handayani S. (2022). The development of real-time polymerase chain reaction for identification of beef meatball. Applied Food Research. 2: 100148. [DOI: 10.1016/j.afres.2022.100148] [DOI:10.1016/j.afres.2022.100148]
99. Roy D., Akhter S., Sarker A., Hossain M., Lyzu C., Mohanta L., Islam D., Khan M. (2022). Tracing the pig and cattle origin in processed food and feed products targeting mitochondrial 12S rRNA gene. Journal of Food Quality and Hazards Control. 9: 163-171. [DOI: 10.18502/jfqhc.8.4.8256] [DOI:10.18502/jfqhc.8.4.8256]
100. Sahilah A.M., Norhayati Y., Norrakiah A.S., Aminah A., Wan Aida W.M. (2011). Halal authentication of raw meats using PCR amplification of mitochondrial DNA. International Food Research Journal. 18: 1489-1491.
101. Sajali N., Wong S.C., Bakar S.A., Mokhtar N.F.K., Manaf Y.N.A., Yuswan M.H., Desa M.N.M. (2020). Analytical approaches of meat authentication in food. International Journal of Food Science and Technology. 56: 1535-1543. [DOI: 10.1111/ ijfs.14797] [DOI:10.1111/ijfs.14797]
102. Sanjaya U.H., Arabella R. (2023). Legal protection of consumer data on e-commerce platforms with cash on delivery (COD) systems. KnE Social Sciences. 8: 20-32. [DOI: 10.18502/kss.v8i1.10869]
103. Satam H., Joshi K., Mangrolia U., Waghoo S., Zaidi G., Rawool S., Thakare R.P., Banday S., Mishra A., Das G., Malonia S.K. (2023). Next-generation sequencing technology: current trends and advancements. Biology.12: 997. [DOI: 10.3390/biology12070997] [DOI:10.3390/biology12070997] [PMID] [PMCID]
104. Schloss P.D., Gevers D., Westcott S.L. (2011). Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLOS ONE. 6: e27310. [DOI: 10.1371/journal.pone.0027310] [DOI:10.1371/journal.pone.0027310] [PMID] [PMCID]
105. Sedlazeck F.J., Rescheneder P., Smolka M., Han F., Nattestad M., Von Haeseler A., Schatz M.C. (2018).Accurate detection of complex structural variations using single-molecule sequencing. Nature Methods. 15: 461-468. [DOI: 10.1038/s41592-018-0001-7] [DOI:10.1038/s41592-018-0001-7] [PMID] [PMCID]
106. Segata N., Waldron L., Ballarini A., Narasimhan V., Jousson O., Huttenhower C. (2012). Metagenomic microbial community profiling using unique clade-specific marker genes. Nature Methods. 9: 811-814. [DOI: 10.1038/nmeth.2066] [DOI:10.1038/nmeth.2066] [PMID] [PMCID]
107. Shaharuddin F.Z., Norman A.A., Hamid S., Zakaria Z. (2025). Halal food industry in Malaysia: issues and challenges. Food Research. 9: 33-42. [DOI: 10.26656/fr.2017.9(s2).072] [DOI:10.26656/fr.2017.9(S2).072]
108. Shen W., Song Z., Zhong X., Huang M., Shen D., Gao P., Qian X., Wang M., He X., Wang T., Li S., Song X. (2022). Sangerbox: a comprehensive, interaction‐friendly clinical bioinformatics analysis platform. iMeta. 1: 15. [DOI: 10.1002/imt2.15] [DOI:10.1002/imt2.15] [PMID] [PMCID]
109. Shirazi S., Meyer R.S., Shapiro B. (2021). Revisiting the effect of PCR replication and sequencing depth on biodiversity metrics in environmental DNA metabarcoding. Ecology and Evolution. 11: 15766-15779. [DOI: 10.1002/ece3.8239] [DOI:10.1002/ece3.8239] [PMID] [PMCID]
110. Slatko B.E., Gardner A.F., Ausubel F.M. (2018).Overview of next-generation sequencing technologies. Current Protocols in Molecular Biology. 122: e59. [DOI: 10.1002/cpmb.59] [DOI:10.1002/cpmb.59] [PMID] [PMCID]
111. Sun K., Liu Y., Zhou X., Yin C., Zhang P., Yang Q., Mao L., Shentu X., Yu X. (2022). Nanopore sequencing technology and its application in plant virus diagnostics. Frontiers in Microbiology. 13: 939666. [DOI: 10.3389/fmicb.2022.939666] [DOI:10.3389/fmicb.2022.939666] [PMID] [PMCID]
112. Taber K.S. (2017). The use of Cronbach's Alpha when developing and reporting research instruments in science education. Research in Science Education. 48: 1273-1296. [DOI: 10.1007/s11165-016-9602-2] [DOI:10.1007/s11165-016-9602-2]
113. Talib M.S.A., Zulfakar M.H. (2023). Sustainable halal food supply chain management in a small rentier halal market. Arab Gulf Journal of Scientific Research. 41: 61-72. [DOI: 10.1108/agjsr-11-2022-0251] [DOI:10.1108/AGJSR-11-2022-0251]
114. Teng J.L.L., Yeung M.L., Chan E., Jia L., Lin C.H., Huang Y., Tse H., Wong S.S.Y., Sham P.C., Lau S.K.P., Woo P.C.Y. (2017). PacBio but not Illumina technology can achieve fast, accurate and complete closure of the high GC, complex Burkholderiapseudomallei two-chromosome genome. Frontiers in Microbiology. 8: 1448. [DOI: 10.3389/fmicb.2017.01448] [DOI:10.3389/fmicb.2017.01448] [PMID] [PMCID]
115. Thomas G.W.C., Hahn M.W. (2019). Referee: reference assembly quality scores. Genome Biology and Evolution. 11: 1483-1486. [DOI: 10.1093/ gbe/evz088] [DOI:10.1093/gbe/evz088] [PMID] [PMCID]
116. Tyler A.D., Mataseje L., Urfano C., Schmidt L., Antonation K., Mulvey M.R., Corbett C.R. (2018). Evaluation of Oxford Nanopore'sMinION sequencing device for microbial whole genome sequencing applications. Scientific Reports. 8: 10931. [DOI: 10.1038/s41598-018-29334-5] [DOI:10.1038/s41598-018-29334-5] [PMID] [PMCID]
117. Udaondo Z., Sittikankaew K., Uengwetwanit T., Wongsurawat T., Sonthirod C., Jenjaroenpun P., Pootakham W., Karoonuthaisiri N., Nookaew I. (2021). Comparative analysis of PacBio and Oxford Nanopore sequencing technologies for transcriptomic landscape identification of Penaeusmonodon. Life. 11: 862. [DOI: 10.3390/life11080862] [DOI:10.3390/life11080862] [PMID] [PMCID]
118. Uliano-Silva M., Ferreira J.G.R.N., Krasheninnikova K., Blaxter M., Mieszkowska N., Hall N., Holland P., Durbin R., Richards T., Kersey P., Hollingsworth P., Wilson W., Twyford A., Gaya E., Lawniczak M., Lewis O., Broad G., Martin F., Hart M., McCarthy S.A. (2023). MitoHiFi: a python pipeline for mitochondrial genome assembly from PacBio high fidelity reads. BMC Bioinformatics. 24: 288. [DOI: 10.1186/s12859-023-05385-y] [DOI:10.1186/s12859-023-05385-y] [PMID] [PMCID]
119. Van Der Spiegel M., Van Der Fels-Klerx H., Sterrenburg P., Van Ruth S.M., Scholtens-Toma I., Kok E. (2012). Halal assurance in food supply chains: verification of halal certificates using audits and laboratory analysis. Trends in Food Science and Technology. 27: 109-119. [DOI: 10.1016/j.tifs.2012.04.005] [DOI:10.1016/j.tifs.2012.04.005]
120. Venkatas J., Adeleke M.A. (2019). Emerging threat of Eimeria operational taxonomic units (OTUs) on poultry production. Parasitology. 146: 1615-1619. [DOI: 10.1017/s0031182019001100] [DOI:10.1017/S0031182019001100] [PMID]
121. Wang H., Chen Y., Wang L., Liu Q., Yang S., Wang C. (2023). Advancing herbal medicine: enhancing product quality and safety through robust quality control practices. Frontiers in Pharmacology. 14: 1265178. [DOI: 10.3389/fphar.2023.1263813] [DOI:10.3389/fphar.2023.1265178] [PMID] [PMCID]
122. Wang X., Cai Y., Sun Y., Knight R., Mai V. (2011). Secondary structure information does not improve OTU assignment for partial 16S rRNA sequences. The ISME Journal. 6: 1277-1280. [DOI: 10.1038/ismej.2011.187] [DOI:10.1038/ismej.2011.187] [PMID] [PMCID]
123. Wasswa F.B., Kassaza K., Nielsen K., Bazira J. (2022). MinION whole-genome sequencing in resource-limited settings: challenges and opportunities. Current Clinical Microbiology Reports. 9: 52-59. [DOI: 10.1007/s40588-022-00183-1] [DOI:10.1007/s40588-022-00183-1] [PMID] [PMCID]
124. Wenger A., Peluso P., Rowell W., Chang P.C., Hall R., Concepcion G., Hunkapiller M. (2019). Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nature Biotechnology. 37: 1155-1162. [DOI: 10.1038/s41587-019-0217-9] [DOI:10.1038/s41587-019-0217-9] [PMID] [PMCID]
125. Wick R.R., Judd L.M., Holt K.E. (2023). Assembling the perfect bacterial genome using Oxford Nanopore and Illumina sequencing. PLOS Computational Biology. 19: e1010905. [DOI: 10.1371/journal.pcbi.1010905] [DOI:10.1371/journal.pcbi.1010905] [PMID] [PMCID]
126. Woltyńska A., Gawor J., Olech M.A., Górniak D., Grzesiak J. (2023). Bacterial communities of Antarctic lichens explored by gDNA and cDNA 16S rRNA gene amplicon sequencing. FEMS Microbiology Ecology. 99: fiad015. [DOI: 10.1093/femsec/fiad015] [DOI:10.1093/femsec/fiad015] [PMID] [PMCID]
127. Woo C., Kumari P., Eo K., Lee W., Kimura J., Yamamoto N. (2023). Combining vertebrate mitochondrial 12S rRNA gene sequencing and shotgun metagenomic sequencing to investigate the diet of the leopard cat (Prionailurusbengalensis) in Korea. PLOS ONE. 18: e0278941. [DOI: 10.1371/journal.pone.0278941] [DOI:10.1371/journal.pone.0278941] [PMID] [PMCID]
128. Wood D.E., Lu J., Langmead B. (2019). Improved metagenomic analysis with Kraken 2. Genome Biology. 20: 257. [DOI: 10.1186/s13059-019-1891-0] [DOI:10.1186/s13059-019-1891-0] [PMID] [PMCID]
129. Yakin A.U. (2021). Halal certification, standards, and their ramifications in Belgium. Islamic Studies Journal. 27: 45-59. [DOI: 10.1163/9789004459236_ 008] [DOI:10.1163/9789004459236_008]
130. Yang Y., Xie B., Yan J. (2014). Application of next-generation sequencing technology in forensic science. Genomics, Proteomics and Bioinformatics. 12: 190-197. [DOI: 10.1016/j.gpb.2014.09.001] [DOI:10.1016/j.gpb.2014.09.001] [PMID] [PMCID]
131. Yang L., Tan Z., Wang D., Xue L. Guan M., Huang T., Li R. (2014). Species identification through mitochondrial rRNA genetic analysis. Scientific Reports. 4: 4089. [DOI: 10.1038/srep04089] [DOI:10.1038/srep04089] [PMID] [PMCID]
132. Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., Madden T.L. (2012). Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics.13: 134. [DOI: 10.1186/ 1471-2105-13-134] [DOI:10.1186/1471-2105-13-134] [PMID] [PMCID]
133. Zhang Y., Qu Q., Mingzhen R., Nana Z., Zhao Y., Tao F. (2020). Simultaneous identification of animal-derived components in meats using high-throughput sequencing in combination with a custom-built mitochondrial genome database. Scientific Reports.10: 8965. [DOI: 10.1038/s41598-020-65724-4] [DOI:10.1038/s41598-020-65724-4] [PMID] [PMCID]
134. Zhou W., Liu X., Lv M., Shi Y., Zhang L. (2023). The recognition mode between hsRBFA and mitoribosome 12S rRNA during mitoribosomal biogenesis. Nucleic Acids Research. 51: 1353-1363. [DOI: 10.1093/ nar/gkac1234] [DOI:10.1093/nar/gkac1234]
135. Zou A., Phan L., Chen S., Campbell J., Guo P., Ren R., Hendrycks D. (2023). Representation engineering: a top-down approach to AI transparency. arXiv. [DOI: 10.48550/arXiv.2301.01912] [DOI:10.48550/arXiv.2301.01912]
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |